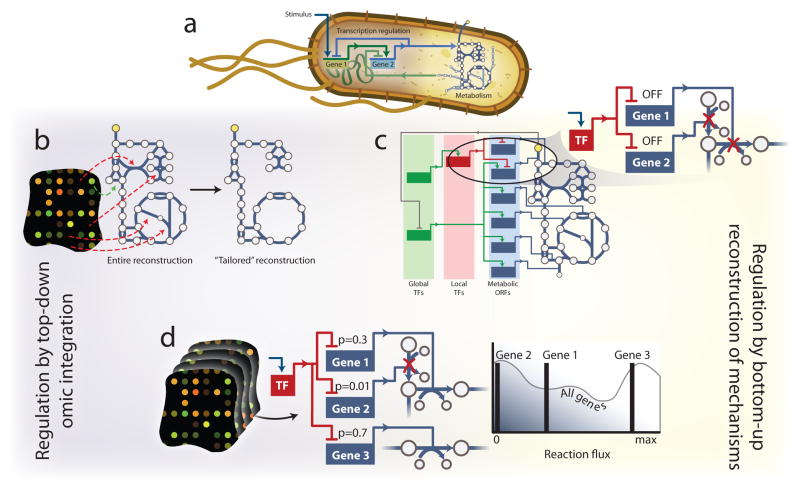
Figure 6. Incorporating and inferring regulation.
(a) Signaling, transcription regulation, and metabolism are interlinked in the cell. Therefore integrating the networks may provide more holistic modeling of organisms. Two primary paradigms exist in COBRA modeling for integrating transcription regulation and metabolism. (b) Algorithms such as GIMME105 and MBA107 use high-throughput data and model simulations to identify which pathways are likely expressed and active in the cells when the data were sampled. This results in a tailored context-specific representation of the metabolic network. (c) Algorithms such as rFBA109, iFBA111, and SR-FBA110 incorporate detailed mathematical representations of the known molecular mechanisms of transcription regulation. These approaches contain binary regulatory logic that dictates, under a specific signal, which metabolic pathways are suppressed and cannot carry flux. (d) Hybrid methods, such as PROM116 are arising, in which transcriptomic data are used to infer the regulatory network. This allows for the elucidation of novel regulatory interactions and their immediate incorporation into model simulations. PROM also uses probabilistic measures to allow for a more continuous regulation of reaction flux. For example, Gene 2 is tightly regulated by a transcription factor (TF). Thus, when the TF is activated by a signal, reaction flux is more tightly constrained than Gene 1, which is only loosely regulated.