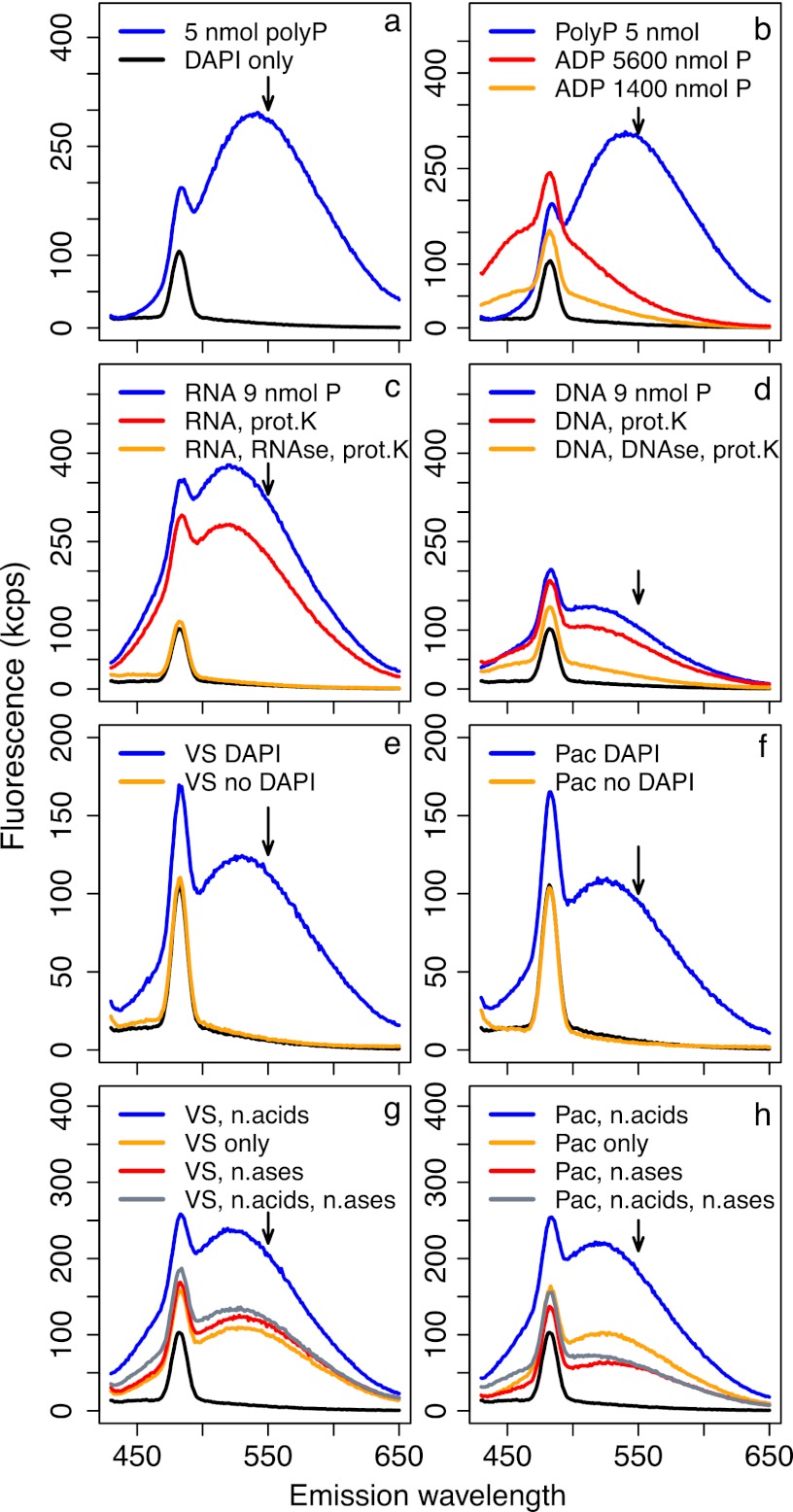
Fig 6.
Fluorescence emission spectra under 415-nm excitation; each spectrum corresponds to a single sample. All solutions were stained with DAPI unless otherwise indicated. Arrows indicate the wavelength at which emission is measured for polyP quantification (550 nm), and black lines in all panels show spectra of blank Tris buffer stained with DAPI. Fluorescence of 5 nmol polyP solutions greatly exceeded that of even very concentrated ADP solutions (a and b). However, 3 μg of RNA and DNA (∼9 nmol P) yielded high fluorescence when stained with DAPI (c and d), which was only partly reduced during our core polyP extraction protocol of boiling and proteinase K (prot.K) treatment (red lines) but almost completely reduced when nuclease treatments were incorporated into the extraction protocol (orange lines). Natural particle samples from the Vineyard Sound (e) and North Pacific (f) showed very clean fluorescence spectra with and without DAPI; stained spectra were consistent with polyP-DAPI fluorescence. When Vineyard Sound (g) and North Pacific (h) samples were amended with 1.5 μg each of DNA and RNA (∼4.5 nmol P), fluorescence was strongly increased relative to that of unamended samples (blue versus orange lines, respectively). This increase was completely removed by treatment with DNase and RNase (gray lines). Treatment with DNase and RNase of samples not amended with nucleic acids (n,acids) did not change fluorescence in Vineyard Sound samples (g, red line) but caused a strong decrease in the North Pacific sample (h, red line).