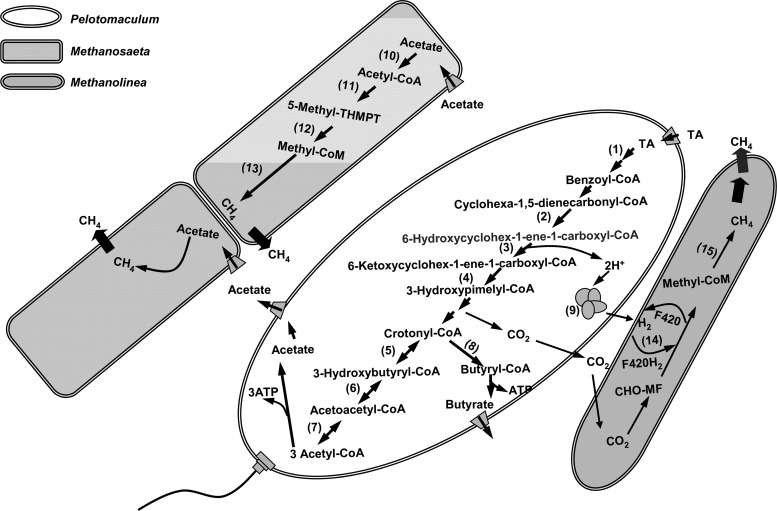
Fig 5.
Metabolic pathways for TA degradation in the TA-degrading Pelotomaculum spp., and for methane formation in acetoclastic Methanosaeta spp. and hydrogenotrophic Methanolinea spp. (Modified from Lykidis et al. [5].) The proteins detected by the proteomic analysis are indicated (by numbers in parentheses in the figure) as follows: 1, UbiD family decarboxylases (tadcc27178); 2, cyclohexa-1,5-dienecarbonyl-CoA hydratase (tadcc30311); 3, 6-hydroxycyclohex-1-ene-1-carboxyl-CoA dehydrogenase (tadcc30312); 4, 6-oxo-cyclohex-1-ene-carbonyl-CoA hydrolase (tadcc30310); 5, crotonase (tadcc30320); 6, β-hydroxybutyryl-CoA dehydrogenase (tadcc30319); 7, acetyl-CoA acetyltransferase (tadcc30308 and tadcc30318); 8, butyryl-CoA dehydrogenase (tadcc30313); 9, Fe-only hydrogenase (tadcc12813); 10, acetyl-CoA synthetase (tadcc27522, tadcc27524, and tadcc32283); 11, acetyl-CoA decarbonylase/synthase (tadcc17916 and tadcc8403); 12, tetrahydromethanopterin S-methyltransferase (tadcc28098 and tadcc28101); 13, methyl-CoM reductase alpha subunit (tadcc11668 and tadcc5364), beta subunit (tadcc6056, tadcc6057, and tadcc5367), and gamma subunit (tadcc6054 and tadcc5365); 14, F420-reducing hydrogenase (tadcc3700); and 15, methyl-CoM reductase beta subunit (tadcc3596) and gamma subunit (tadcc3599).