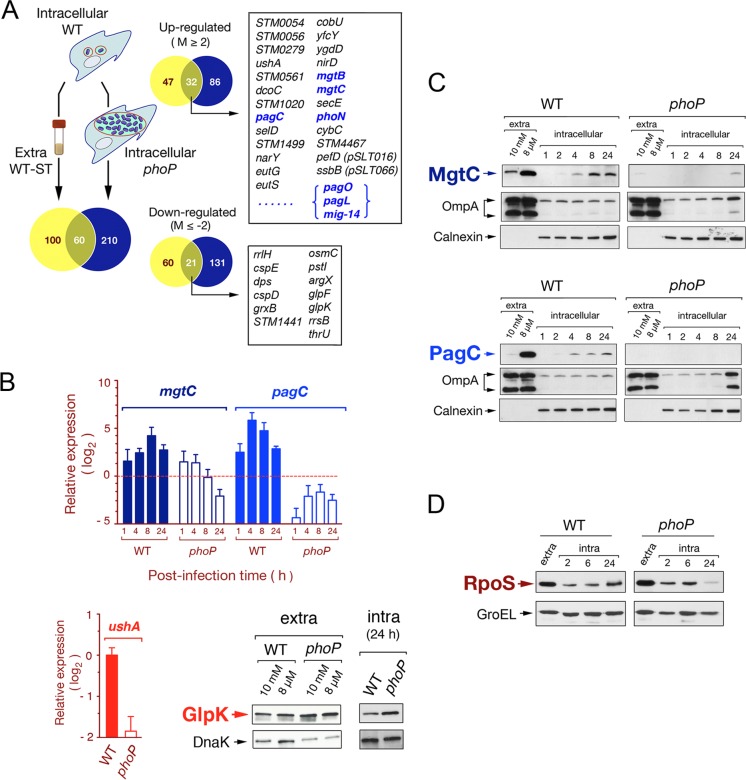
Fig 3.
Characterization of the S. Typhimurium PhoP-PhoQ regulon in dormant nongrowing intracellular bacteria. (A) Venn diagram showing the overlapping among transcriptomes obtained from intracellular nongrowing bacteria (wild-type strain SV5015), intracellular proliferating bacteria (phoP mutant strain MD1120), and extracellular wild-type bacteria grown to stationary phase in LB medium. Each of these transcriptomes refers to the expression of extracellular bacteria grown to exponential phase (see Table S2 in the supplemental material). Numbers of genes differing in expression among transcriptomes by more than 4-fold are indicated. Highlighted in boxes are some of the genes displaying differential expression among transcriptomes (intra-WT versus extra-WT and intra-WT versus intra-phoP), which respond to both the intracellular environment and the functional status of the PhoP-PhoQ system. Genes previously reported to be regulated by PhoP-PhoQ are indicated in blue (see Tables S6 and S7 for details). (B) Validation data obtained by RT-qPCR for two PhoP-PhoQ-regulated genes, mgtC and pagC. Two other genes hitherto not assigned to the PhoP-PhoQ regulon, ushA and glpK, were also validated at the transcript and protein levels, respectively. mgtC and pagC data refer to different postinfection times (1, 4, 8, and 24 h) upon invasion of NRK-49F fibroblasts and are relative to expression levels detected in extracellular bacteria growing to exponential phase. ushA data are relative to the expression levels registered at 24 h postinfection in nongrowing intracellular bacteria. In the RT-qPCR assays, expression values were normalized to those obtained for the ompA gene used as an internal control. (C) Relative levels of the MgtC-3×FLAG- and PagC-3×FLAG-tagged proteins detected in intracellular nongrowing wild-type bacteria and in the overgrowing phoP mutant at the indicated postinfection times. As a control of canonical PhoP-PhoQ regulation, these two proteins were also monitored in extracellular bacteria grown in inducing (8 μM Mg2+) or repressing (10 mM Mg2+) conditions. (D) Levels of the alternative sigma factor RpoS detected in intracellular bacteria at different postinfection times upon entry into NRK-49F fibroblasts. DnaK, OmpA (bacterial proteins), and calnexin (eukaryotic protein) were used as loading controls.