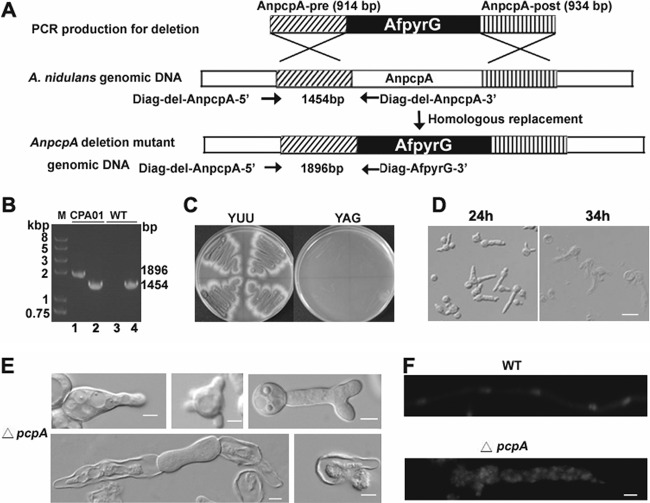
Fig 2.
Identification and analysis of the essential A. nidulans gene AnpcpA using the heterokaryon rescue technique. (A) Diagram showing the deletion strategy for AnpcpA. (B) PCR analysis showing the integration of the AfpyrG nutritional marker into the genome at the original AnpcpA locus in the CPA01 strain while AnpcpA can still be detected, suggesting that transformants were heterokaryons. For lanes 2 and 4, the primers Diag-del-AnpcpA-5′ and Diag-del-AnpcpA-3′ were used to determine whether AnpcpA still exists in the genome, and the expected size is 1,454 bp. For lanes 1 and 3, primers Diag-del-AnpcpA-5′ and Diag-AfpyrG-3′ were used to determine whether there was a homologous recombination to replace AnpcpA with the nutritional marker gene pyrG in the genome, and the expected size is 1,896 bp. In lanes 3 and 4, genomic DNA of TN02A7 was used as a PCR template; for lanes 1 and 2, genomic DNA of transformants was used as a PCR template. (C) Replica transformant spores from heterokaryons on YAG and YUU. (D) Conidial spores from heterokaryon transformants cultured in liquid YAG for 24 h, showing two kinds of genotypes in the progeny: ungerminated spores were pcpA wild type in the pyrG mutation background, and the other germlings had the pcpA deletion. Bar, 10 μm. (E) Phenotypes of abnormal germlings in progeny of AnpcpA deletion transformants. (F) Germlings from progeny of AnpcpA deletion transformants were stained with DAPI, showing the abnormal distribution of nuclei. Bar, 5 μm.