Fig 1.
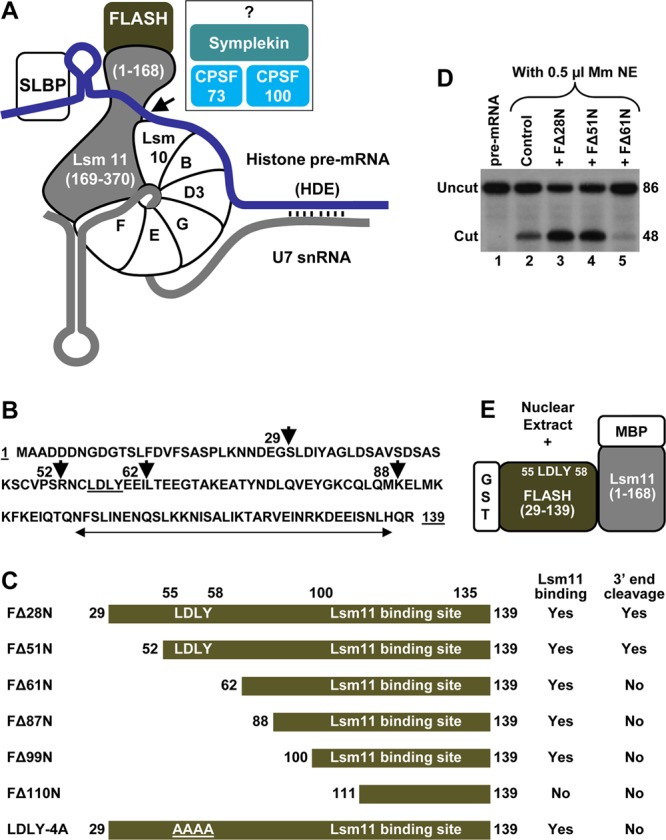
Biochemical assay to identify proteins interacting with a complex of FLASH and Lsm11. (A) The known critical factors in 3′-end processing of histone pre-mRNA (blue line) are shown. Vertical lines indicate base paring between the histone downstream element (HDE) and the 5′ end of U7 snRNA (gray line). Histone pre-mRNAs are cleaved by CPSF73 typically five nucleotides after the stem-loop (arrow). The reaction also requires CPSF100, symplekin, and potentially other factors (question mark). (B) Amino acid sequence of the N-terminal region of human FLASH (amino acids 1 to 139). The first amino acids of various N-terminal truncations are indicated with arrowheads. The Lsm11-binding site is indicated with a double-headed arrow, and the LDLY motif is underlined. (C) Diagram of FLASH mutants used in this study. The GST tag is fused N terminally to each protein and is not indicated. The ability of each FLASH protein to support processing or bind Lsm11 is indicated. (D) In vitro processing of histone pre-mRNA with a limiting amount of mouse nuclear extract (NE) in the absence of any recombinant protein (lane 2) or in the presence of 100 ng of various FLASH proteins, as indicated (lanes 3 to 5). Lane 1 contains the input substrate. (E) Schematic of the assay for isolation of factors that bind to the FLASH/Lsm11 complex. The N-terminal FLASH (amino acids 29 to 139) fused to GST and the N-terminal Lsm11 (amino acids 1 to 168) fused to MBP were incubated with a nuclear extract, and interacting proteins were purified on glutathione beads.
