Fig 6.
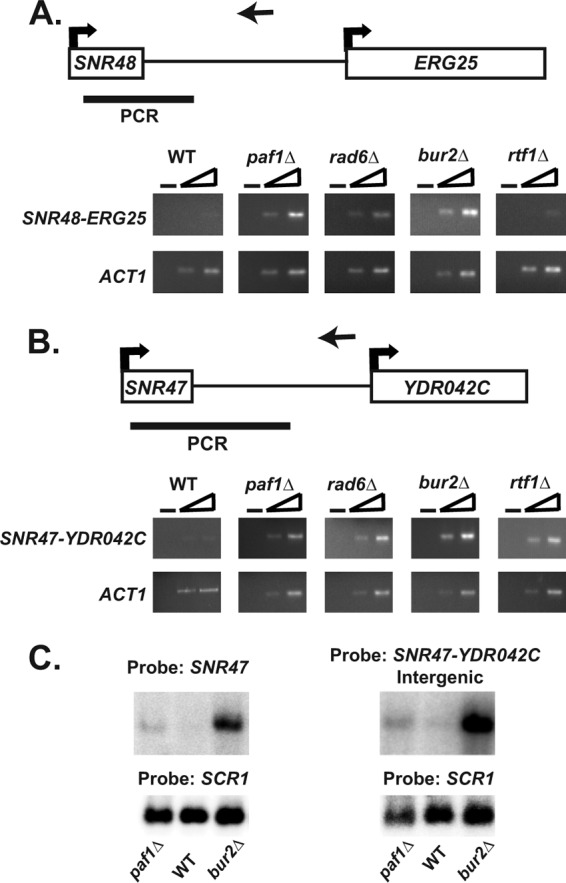
Analysis of SNR47 and SNR48 read-through transcripts. (A and B) As described in Materials and Methods, strand-specific cDNA was generated using a primer downstream of either SNR47 or SNR48 (primer locations indicated by the arrows pointing left). For a control, a no-reverse transcriptase (no-RT) reaction was performed on each sample as indicated by the minus sign. PCRs were done with two different concentrations of each cDNA using a primer within the snoRNA sequence and a primer downstream of the snoRNA (sequences found in PCR product indicated by a black bar). ACT1 is used as a positive control for RT-PCR experiments. RT-PCR analysis was performed in triplicate from independent biological replicates. See Table 2 for the sequences of the primers used. (A) Representative RT-PCRs at SNR48 from wild-type (KY1699), paf1Δ (KY1700), rad6Δ (KY2045), bur2Δ (KY1718), and rtf1Δ (KY1703) cells. (B) Representative RT-PCRs at SNR47 from wild-type (KY1699), paf1Δ (KY1700), rad6Δ (KY2339), bur2Δ (KY1718), and rtf1Δ (KY1703) cells. (C) Northern blot analysis using RNA from wild-type (KY2276), paf1Δ (KY1702), and bur2Δ (KY2409) cells. Identical RNA was run on the same gel in parallel, and then after the gel was transferred to a membrane, each half of the membrane was hybridized with either a probe against the snoRNA sequence of SNR47 or a probe against the regions downstream of SNR47 (SNR47-YDR042C). Both probes recognized the SNR47 read-through transcript depicted.
