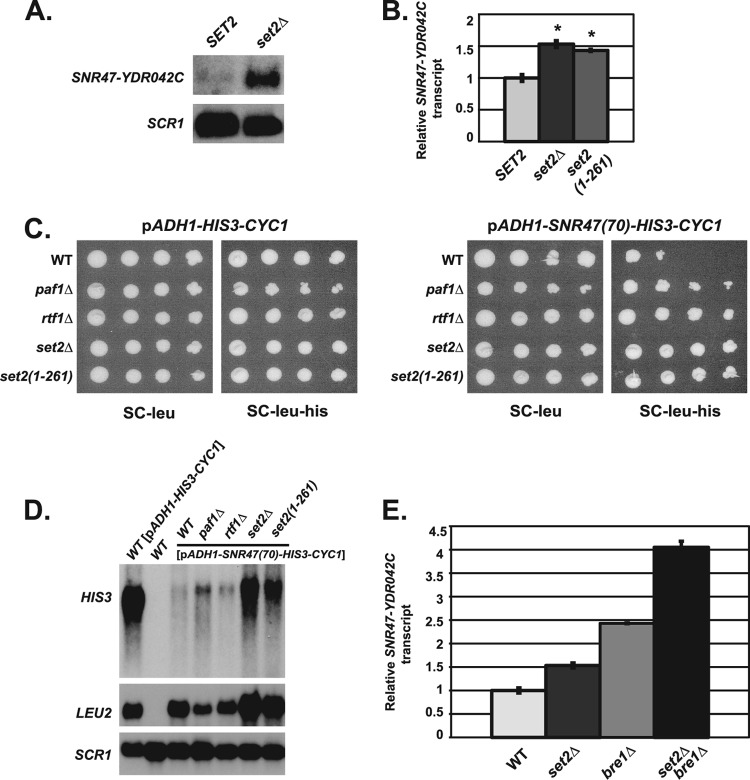
Fig 7.
Set2 is involved in snoRNA termination through a pathway distinct from H2B ubiquitylation. (A) Representative Northern blot analysis of extended SNR47 transcripts (SNR47-YDR042C) in wild-type (KY2090) and set2Δ (KY2280) cells. SCR1 transcript levels serve as a loading control. (B) Quantification of SNR47-YDR042C transcript levels in wild-type (KY2048), set2Δ (KY2282), and set2(1-261) (KY2281) cells measured by RT-qPCR, with the relative signal of the wild type set at 1. The SEMs are indicated by the error bars. Asterisks indicate a P value of <0.0025 relative to wild type. (C) The following his3Δ200 leu2Δ1 strains were transformed with an established LEU2-marked SNR47 reporter plasmid [pADH1-SNR47(70)-HIS3-CYC1] or control plasmid (pADH1-HIS3-CYC1) (see Materials and Methods for more information): wild-type (KY2042), paf1Δ (KY1664), rtf1Δ (KY2041), set2Δ (KY2280), and set2(1-261) (KY2338). Tenfold serial dilutions of cells containing the listed plasmids were spotted on synthetic complete medium lacking leucine (SC-leu) and synthetic complete medium lacking leucine and histidine (SC-leu-his) and incubated for 5 days at 30°C. (D) Representative Northern blot analysis examining HIS3 transcripts from the SNR47 reporter plasmid in his3Δ200 leu2Δ1 strains (as in panel C). Wild-type cells with the control plasmid (pADH1-HIS3-CYC1) show the size of the HIS3 transcript lacking 70 bp of the SNR47 sequence. LEU2 transcript levels serve as a control for plasmid levels, and SCR1 transcript levels control for total RNA. RNA from a his3Δ200 leu2Δ1 strain (KY2090) containing no plasmids (WT) (lane 2) serves as a control for probe specificity. (E) Quantification of SNR47 extended transcripts (SNR47-YDR042C) in wild-type (KY2048), set2Δ (KY2282), bre1Δ (KY2046), and set2Δ bre1Δ (KY2283) cells determined by RT-qPCR, with the relative signal of the wild type set at 1. The SEMs are indicated by the error bars.