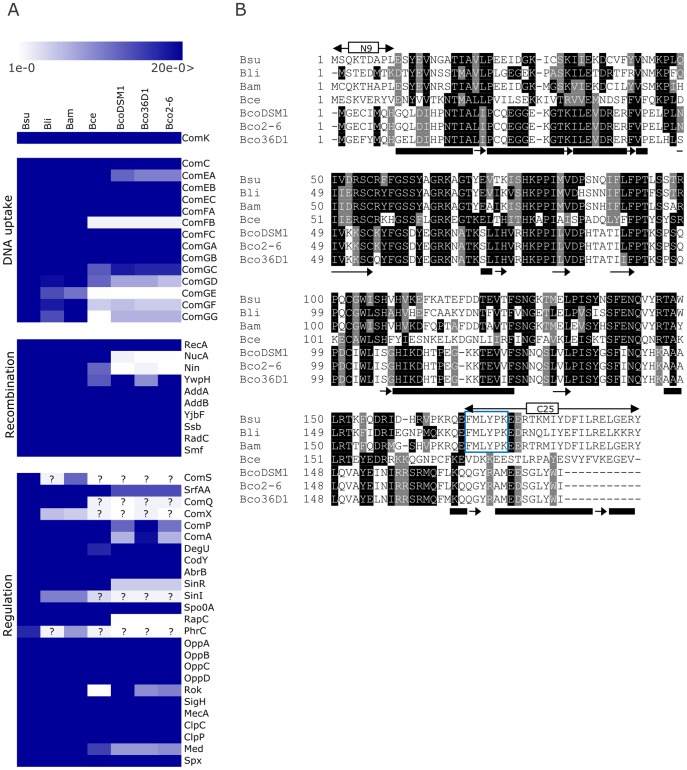
Figure 1. Survey on the presence of competence genes and the alignment of ComK protein sequences from various Bacillus strains.
(A) Results of BLAST searches were visualized with Genesis 1.6 software: white is absent (with E-value of E–0), dark blue is present (E-value<E–20). BLAST analysis was performed with B. subtilis protein sequences against translated protein database of a given genome. Protein names are indicated on the right. Bsu, B. subtilis; Bli, B. licheniformis, Bam, B. amyloliquefaciens, Bce, B. cereus Bco, B. coagulans. Question marks denote small ORFs where identification is uncertain using the available bioinformatic tools that can miss homologues. (B) Multiple alignment of ComK homologues. Black background represents conserved amino acids and grey background represents similar amino acids. Alignment was performed using ClustalW [59], and presented using Boxshade 3.21 program. The N- and C-terminal deletions analyzed by Susanna et al [47] are marked (ΔN9 and ΔC25, respectively). Boxed amino acid residues indicate the residues involved in interaction with MecA [60]. Alpha-helices and beta-sheets of B. subtilis ComK protein are indicated with rectangles and arrows under the alignment, respectively.