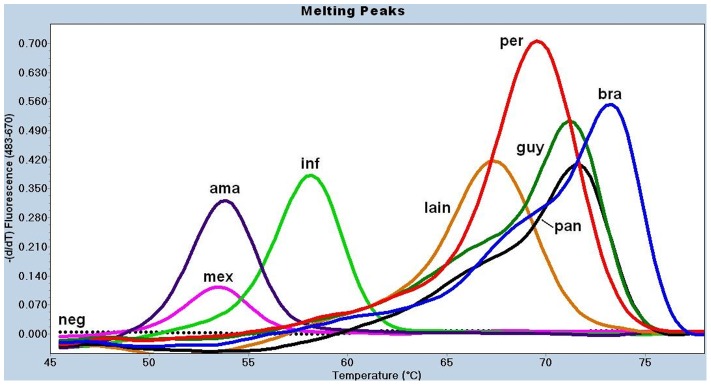
Figure 1. Melting curve analysis for the MPI-based real-time PCR assay on Leishmania reference strains.
Five nanograms of DNA (references) or 5 ul of nested-PCR products (samples) were used as template to run the assay. The species designation is given based on the Tm calculated for each melting peak (point in the x axis for the peak of the curve). FRET probes on L. (L.) amazonensis (ama) or L. (L.) mexicana (mex) DNA are melted at lower temperature (53°C) while on L. (V.) braziliensis (bra) DNA are melted at the highest temperature (74°C). This assay could not differentiate between L. (V.) guyanensis (guy) and L. (V.) panamensis (pan) due to their overlapping Tm (72°C). All assessed Leishmania species yielded a melting curve. A straight dotted line corresponds to the negative controls (neg), DNA free or parasite-free human DNA. inf = L. infantum; lain = L. (V.) lainsoni; per = L. (V.) peruviana.