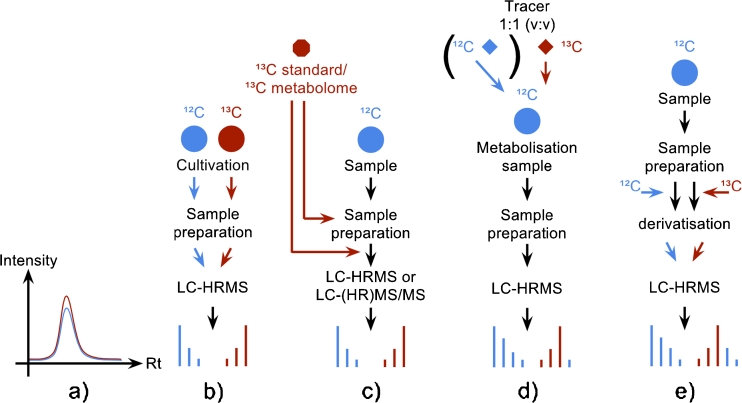
Fig. 1.
Different strategies for using SIL to assist metabolomics studies. a 13C, 15N, and 34S-enriched substances are not chromatographically separated from the corresponding natural isotopologues, thus the non-labeled and the labeled isotopologues elute at the same retention time with identical peak profiles. b For non-targeted annotation of an organism’s metabolome the organism can be cultivated in parallel using differently isotopologue-enriched nutrition sources (e.g., 12C and 13C glucose as sole carbon source). The extracts are subsequently mixed and measured with LC–HRMS. The resulting data pattern helps in the extraction of true biological signals. c Absolute compound quantification using an authentic, labeled standard or relative quantification using a stock of globally labeled sample extract of the same organism for inter-experiment comparison. d Metabolism experiment using natural and fully labeled tracer substances enables metabolism studies and greatly helps to separate products of metabolism from other biological signals. In contrast with metabolism studies, fluxomics experiments only spike with the labeled tracer. e Derivatisation using non-labeled and labeled derivatisation agents enables rapid recovery of many metabolites belonging to the same chemical groups (e.g., alcohols, acids …)