Background: Nm23-H4 is a mitochondrial nucleoside diphosphate kinase that binds mitochondrial membranes.
Results: Nm23-H4 interaction with GTPase OPA1 provides a local GTP supply; its interaction with anionic phospholipids inhibits kinase activity but allows intermembrane cardiolipin transfer and sensitizes for apoptosis.
Conclusion: Nm23-H4 is a bifunctional switch operated by cardiolipin.
Significance: The cardiolipin transfer property has various implications, e.g. for lipid metabolism and apoptosis.
Keywords: Bioenergetics, Cardiolipin, Lipids, Mitochondrial Apoptosis, Molecular Modeling, Lipid Trafficking, Lipidomics, Mass Spectrometry, Nucleotides, Protein-Lipid Interaction
Abstract
The nucleoside diphosphate kinase Nm23-H4/NDPK-D forms symmetrical hexameric complexes in the mitochondrial intermembrane space with phosphotransfer activity using mitochondrial ATP to regenerate nucleoside triphosphates. We demonstrate the complex formation between Nm23-H4 and mitochondrial GTPase OPA1 in rat liver, suggesting its involvement in local and direct GTP delivery. Similar to OPA1, Nm23-H4 is further known to strongly bind in vitro to anionic phospholipids, mainly cardiolipin, and in vivo to the inner mitochondrial membrane. We show here that such protein-lipid complexes inhibit nucleoside diphosphate kinase activity but are necessary for another function of Nm23-H4, selective intermembrane lipid transfer. Mitochondrial lipid distribution was analyzed by liquid chromatography-mass spectrometry using HeLa cells expressing either wild-type Nm23-H4 or a membrane binding-deficient mutant at a site predicted based on molecular modeling to be crucial for cardiolipin binding and transfer mechanism. We found that wild type, but not the mutant enzyme, selectively increased the content of cardiolipin in the outer mitochondrial membrane, but the distribution of other more abundant phospholipids (e.g. phosphatidylcholine) remained unchanged. HeLa cells expressing the wild-type enzyme showed increased accumulation of Bax in mitochondria and were sensitized to rotenone-induced apoptosis as revealed by stimulated release of cytochrome c into the cytosol, elevated caspase 3/7 activity, and increased annexin V binding. Based on these data and molecular modeling, we propose that Nm23-H4 acts as a lipid-dependent mitochondrial switch with dual function in phosphotransfer serving local GTP supply and cardiolipin transfer for apoptotic signaling and putative other functions.
Introduction
In addition to the crucial role in ATP generation and cellular energy homeostasis, mitochondria have been recognized for their other vital functions in various biosynthetic and cell signaling pathways, in particular life/death decisions (1). The identification of these alternative functions of mitochondria has led to new concepts on their likely involvement in the etiology of divergent pathologies such as neurodegenerative, neuromuscular, or cardiovascular diseases, diabetes, and cancer. At the molecular level, several mitochondrial proteins have been identified as being able to acquire dual functions: bioenergetics and apoptotic signaling (2). One of the most prominent examples is cytochrome c that functions as an electron carrier in the respiratory chain but also as cardiolipin (CL)3 oxygenase and an apoptotic trigger once liberated into the cytosol (3, 4). Previously, we have shown that mitochondrial nucleoside diphosphate kinase, NDPK-D, also called nonmetastatic protein 23-H4 (Nm23-H4), not only catalyzes reversible phosphotransfer between Mg2+-complexed NTPs and NDPs (5) but also promotes intermembrane migration of anionic lipids in vitro between vesicle populations, particularly those containing CL (6).
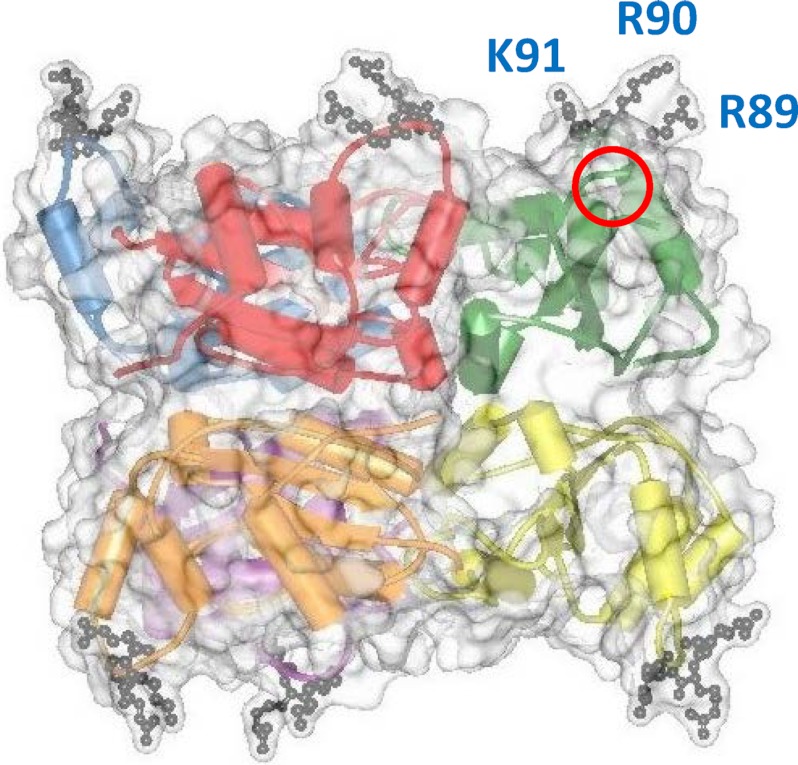
Nm23-H4 has been characterized by our groups in some detail (5–10). It belongs to the Nm23 family comprising 10 isoforms in humans and probably all mammals (genes NME1 to NME10) (11, 12), but Nm23-H4 is the only one with a cleavable N-terminal sequence that targets it into mitochondria. Nm23-H4 forms symmetrical homohexamers with six independent active sites situated at the top and bottom faces of the protein (see red circle in Fig. 1). It is localized in the intermembrane space (IMS) of mitochondria where it is mainly bound to the inner mitochondrial membrane (IM). Although Nm23-H4 catalyzes reversible phosphotransfer between various NDP and NTP species, permanent ATP delivery to IMS via the adenylate translocase (ANT) drives the phosphotransfer reaction toward the generation of other NTPs, principally the most abundant GTP. It has been speculated that this GTP is necessary to sustain local GTPases not satisfied by the import of cytosolic GTP into the IMS (8), such as optic atrophy 1 (OPA1), an essential player in mitochondrial fusion and cristae remodeling (13, 14). The ADP regenerated in the IMS by the Nm23-H4 phosphotransfer reaction was shown to be directly taken up via ANT into the matrix space to stimulate respiratory ATP regeneration (8). This “metabolite channeling” mechanism couples the phosphotransfer reaction to respiration whereby consumed ATP is directly replenished. Binding of Nm23-H4 to IM is important for this channeling mechanism, because it is largely diminished in a membrane binding-deficient Nm24-H4 mutant (see below) (8).
FIGURE 1.
Basic features of the Nm23-H4 structure. The high resolution x-ray structure (PDB code 1EHW) (5, 48) shows the symmetrical oligomer (transparent surface representation) composed of six monomers (in different colors, schematic backbone representation). In one of the six monomers, a red circle indicates the catalytic phosphotransfer site, and blue letters indicate the primary cardiolipin-binding residues (in ball-and-stick representation). Note: residue numbering here and throughout the paper corresponds to nascent, nonprocessed Nm23-H4, whereas numbering in the crystal structure 1EHW corresponds to processed Nm23-H4 that lacks the N-terminal 33 amino acids (mitochondrial targeting peptide).
Binding of Nm23-H4 to the mitochondrial membranes relies on tight complex formation with anionic phospholipids, in particular CL to which it binds with a KD of ∼30 nm (8). The interaction is primarily mediated by a conserved cationic triad (RRK motif) present in each Nm23-H4 monomer (see gray residues and blue letters in Fig. 1). Mutation of the central arginine in the RRK motif (R90D) inhibits a phospholipid interaction yet does not affect the phosphotransfer activity or the oligomeric state of the kinase (8). Because of its symmetrical structure, membrane binding can also occur simultaneously at two opposite faces of the homohexamer (Fig. 1), leading to cross-linking of two membranes in vitro and formation of contact sites between IM and the mitochondrial outer membrane (OM) in vivo. It has long been speculated that such contact sites promote intermembrane lipid transfer (15). Membrane cross-linking by Nm23-H4 in vitro indeed facilitates transfer of lipids between two liposomal membranes without inducing membrane fusion (6). However, the importance of this Nm23-H4 function in vivo, in particular for CL transfer and its potential role in apoptosis, remained unclear.
CL is normally confined to the IM, with its site of biosynthesis being the inner leaflet, and it serves multiple functions within this membrane (16, 17). However, CL can also appear in the OM with numerous biological consequences. It can exchange with mitochondrion-associated endoplasmic reticulum for further CL remodeling or interfere with mitochondrial fusion (16, 17), and it can also play a role in apoptotic signaling (by recruiting pro-apoptotic proteins (18–22)) or mitophagy (3, 23–25). Despite this importance of mitochondrial CL trafficking, the molecular mechanisms involved and their regulation are poorly defined. Few proteins were reported or suspected so far to exchange lipids between mitochondrial membranes, including tBid (26), ATAD3 (27, 28), and Ups1 (29, 30), and to promote the necessary lipid flip-flop between inner and outer membrane leaflets, including proteins like scramblase 3 (31, 32) or Bax (33).
Here, we aimed at delineating phosphotransfer versus lipid transfer activities of Nm23-H4. We show that Nm23-H4 interacts with the following: (i) the mitochondrial GTPase OPA1, suggesting its direct involvement in GTP supply, and (ii) anionic phospholipids leading to inhibition of the phosphotransfer activity. Thus, phosphotransfer and lipid transfer activities are mutually exclusive and regulated by binding with anionic phospholipids (CL). Importantly, ESI-MS-based lipidomics of HeLa mitochondrial subfractions provided evidence for Nm23-H4-mediated intermembrane CL transfer from IM to OM occurring in live cells and sensitizing these cells to apoptosis. These data and molecular modeling suggest that Nm23-H4 acts as a lipid-dependent mitochondrial switch with dual functions in phosphotransfer and CL trafficking.
EXPERIMENTAL PROCEDURES
Materials
Recombinant expression and purification of Nm23-H4 and generation of anti-human Nm23-H4 polyclonal antibodies in rabbits is described elsewhere (5). For immunocytochemistry, Nm23-H4 antibodies were affinity-purified by using recombinant Nm23-H4.
Immunoprecipitation of Mitochondrial Proteins
Mitochondria from male Wistar rat liver were isolated by differential centrifugation as described previously (34); the entire procedure was performed at 4 °C with buffers complemented with protease inhibitors (mixture tablets from Roche Applied Science). For immunoprecipitation (IP), mitochondria (1 mg of protein) were first incubated overnight with Nm23-H4 antibody serum (4 μl) in a total volume (360 μl) of IP buffer (25 mm MOPS, pH 7.0, 50 mm NaCl, 5 mm MgCl2, 10% glycerol, 0.6% BSA, 0.5% Tween) on an orbital shaker. As a control, incubation with preimmune (instead of immune) serum or without serum was carried out in parallel. Protein A-Sepharose beads (Sigma), pre-equilibrated in IP buffer, were added, and the suspension was incubated for 3 h. Beads were washed nine times in IP buffer, the last three times without BSA in the IP buffer. Washed protein-containing beads were directly used for SDS-PAGE and immunoblotting.
Immunoblot Analysis
Proteins attached to antibody-coated Sepharose beads or present in HeLa cells or submitochondrial fractions were separated on 12% SDS-polyacrylamide gels and immunoblotted as described previously (8) using the following primary antibodies: anti-OPA1 (BD Biosciences; 1:500); anti-VDAC total (1:500; a gift of Prof. M. Colombini, University of Maryland); anti-ANT total (Santa Cruz Biotechnology; 1:200); anti-Nm23-H4 (1:7500) and anti-complex III subunit Core 2 (MitoSciences, OR; 1:1000); anti-actin (Sigma; 1:2000); anti-cytochrome c (BD Biosciences; 1:2000); anti-cleaved caspase 3 (Abgent, 1:250), and anti-Bax (Cell Signaling, 1:1000). Blots were revealed with appropriate peroxidase-coupled antibodies and ECL PlusTM substrate (GE Healthcare).
Phosphotransfer Activity
NDPK enzyme activity of Nm24-H4 was determined with a photometric coupled enzyme assay as described previously (5, 35) using kinase buffer (50 mm Tris-HCl, pH 7.4, 50 mm KCl, 5 mm MgCl2, 1 mg/ml BSA, and 100 μm Ap5A to inhibit endogenous adenylate kinase). Briefly, 0.2 mm ATP and 0.2 mm TDP were used as substrates (all substrates from Roche Applied Science), and ADP production was coupled with pyruvate kinase (Sigma) and lactate dehydrogenase (Roche Applied Science) to NADH oxidation. For lipid inhibition experiments, dry lipids (all from Avanti Polar Lipids, Alabaster, AL) were hydrated in TES buffer (10 mm TES, pH 7.0, 50 mm potassium acetate) to a concentration of 5 mg/ml and finally dispersed by vortexing to produce multilamellar vesicles. Phosphotransfer activity assays with pure lipids and purified recombinant Nm23-H4 were carried out by addition of vesicle preparations to a final concentration of 100 μg/ml lipid and, in the case of detergent experiments, by addition of additional 0.5% (v/v) Triton X-100 and 5 min of incubation. Phosphotransfer activity assays with purified intact mitochondria were carried out in isotonic kinase buffer (supplemented with 220 mm mannitol) after 15 min of preincubation of mitochondria (∼30 μg of protein) at 4 °C without or with addition of 0.5% (v/v) Triton X-100.
Cell Culture
HeLa T-RexTM-HeLa clones (Invitrogen) stably expressing Nm23-H4 wild type (WT) or R90D mutants and their growth conditions have been described elsewhere (8). Clones overexpress comparable levels of both proteins already without specific induction. Maximal expression levels of both Nm23-H4 proteins can be achieved by incubation with 1 μg/ml tetracycline for 24 h. Cells were treated for 24 h with 1 μm rotenone to induce apoptosis.
Fluorescence Microscopy and Immunocytochemistry
For mitochondrial staining and Nm23-H4 immunodetection, cells cultivated on microscope glass slides were incubated with 50 nm of the mitochondrion-selective dye MitoTrackerTM Red CMXRos (Molecular Probes) for 30 min at 37 °C, fixed in 3.7% paraformaldehyde, permeabilized in PBS containing 0.5% Triton, blocked in PBS containing 3% BSA, and incubated with anti-Nm23-H4 affinity-purified antibody (dilution 1:500) and finally with the Alexa-Fluor 488-conjugated secondary anti-rabbit antibodies. Slides were mounted with Fluoromount-G (Electron Microscopy Sciences, Hatfield, PA) and examined with a Leica HC microscope.
Isolation of Submitochondrial Fractions from HeLa Cells
Mitochondrial subfractions were isolated from HeLa cells at 4 °C as described previously (8). Briefly, for isolation of mitochondria, HeLa cells were rinsed twice in ice-cold PBS, 1 mm EDTA, scraped into a small volume of ice-cold PBS, 1 mm EDTA, collected by centrifugation (5 min at 700 × g), and resuspended in HEPES/mannitol/sucrose buffer (10 mm HEPES, pH 7.5, 210 mm mannitol, 70 mm sucrose, 1 mm EDTA). Cells were subsequently homogenized by 6 passages through a 25-gauge needle. The homogenate was centrifuged for 5 min at 2000 × g, and the supernatant was centrifuged for another 10 min at 13,000 × g. The crude mitochondrial pellet was resuspended in 10 mm HEPES, pH 7.5, 210 mm mannitol, and 70 mm sucrose, 100 μm EGTA, and washed twice by 10 min of centrifugation at 10,000 × g. Mitochondrial membrane fractions were isolated from this preparation by a swelling-shrinking procedure and ultracentrifugation in a sucrose gradient according to Hovius et al. (36) with some modifications (37). Briefly, the mitochondrial pellet was resuspended in 10 mm KH2PO4 buffer, pH 7.4, and incubated for 15 min under stirring. Then an equal volume of 10 mm KH2PO4, 30% (w/v) sucrose, 30% (v/v) glycerol, 10 mm MgCl2, and 4 mm ATP was added, and the suspension was incubated for another 60 min. The suspension was sonicated with a Branson sonifier (three times for 15 s with 60-s intervals) and centrifuged (12,000 × g for 10 min), and the pellet was resuspended in 100–200 μl of 10 mm KH2PO4, pH 7.4. Both pellet and supernatant were layered separately onto discontinuous sucrose gradients (51, 37, and 25% (w/v) sucrose) and centrifuged at 100,000 × g for 12 h. Fractions at the 25–37 and the 37–51% interphase were collected, containing light OM and heavier IM, respectively.
Mass Spectrometry
Lipids were extracted from IM and OM by the Folch procedure (38). Identification, quantitation, and structural analysis of CL was performed using a Dionex UltimateTM 3000 HPLC coupled on-line to ESI and a linear ion trap mass spectrometer (LXQ Thermo-Fisher) as described previously (39). Identification and quantitation were confirmed on a Q-Exactive instrument (Thermo, Inc., Sunnyvale, CA) and on a Waters Q-TOF Premier instrument (Waters, Inc. Milford, MA), respectively. Quantitation of CL was performed on a Waters Q-TOF mass spectrometer coupled to a Shimadzu Prominence HPLC system (Shimadzu, Inc., Kyoto, Japan). Separation of individual phospholipid classes was accomplished by normal phase chromatography using a silica column (Luna, 3 μm, 15 cm × 2 mm inner diameter, Phenomenex, Inc., Torrance, CA). Phospholipids were eluted from the column using the following solvent systems: solvent A: chloroform, methanol, 30% ammonium hydroxide (80:19.5:0.5, v/v/v); solvent B: chloroform, methanol, water, 30% ammonium hydroxide, (60:34:5.5:0.5). Flow was maintained at 0.2 ml/min, and a linear gradient of 0–100% solvent B in 20 min was run. Separation was performed at room temperature. Parameters for the Q-TOF were as follows: capillary voltage, 2.85 kV, negative mode; source temperature, 100 °C; desolvation gas, 400 liters/h; sampling cone, 60 V; extraction cone, 4.5 V; ion guide, 2.5 V. Tuning was optimized for all lipids across the scan range. For quantitative analysis of CL, samples were mixed with internal standard (1,1′,2,2′-tetramyristoyl-CL, Avanti Polar Lipids) and compared with standard curves of tetralinoleoyl-CL (Avanti Polar Lipids) containing a fixed amount of 1,1′,2,2′-tetramyristoyl-CL internal standard. The amounts of individual molecular species of CL were calculated by determining the ratio of the peak area of the CL molecular species of interest to the internal standard. The CL content was expressed in picomoles of CL per μg of protein in the relevant membrane fraction.
Identification of CL species was also performed on a Q-Exactive mass spectrometer (Thermo) coupled to a Thermo Ultimate 3000 HPLC system. Separation of CL from the remaining phospholipids was accomplished by reverse phase chromatography using a C18 column (Luna, 3 μm, 15 cm × 2 mm inner diameter, Phenomenex). Lipids were eluted using an isocratic system consisting of propanol/water/triethylamine/acetic acid (450:50:2.5:2.5). Flow was maintained at 0.2 ml/min. Parameters for the Q-Exactive were as follows: capillary voltage, 4.0 kV, negative mode; source temperature, 250 °C. Resolution was set to 140,000. Tuning was optimized for all lipids across the scan range.
Apoptosis Assays
Annexin V kit (Biovision, Milpitas, CA) was used to measure externalization of phosphatidylserine. In brief, harvested cells were stained with annexin V-fluorescein isothiocyanate and propidium iodide for 5 min in the dark before flow cytometry analysis. Cell debris as represented by distinct low forward and side scatter were gated out for analysis. Ten thousand events were collected on a FACScan flow cytometer (BD Biosciences) supplied with CellQuest software. Caspase-3/7 activity was measured using a luminescence Caspase-GloTM3/7 assay kit (Promega, Madison, WI) according to the manufacturer's instructions. For cytochrome c, cells were harvested and resuspended in lysis buffer A (250 mm sucrose, 20 mm HEPES-KOH, pH 7.5, 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin) containing 0.05% digitonin for 4 min on ice and then centrifuged at 10,000 × g for 10 min. The resulting supernatants were collected as cytosolic fractions for evaluation of cytochrome c release from mitochondria by immunoblotting.
Molecular Modeling
The orientation of Nm23-H4 (PDB code 1EHW) with respect to the membrane was predicted using the Peripheral Proteins in Membranes (PPM) server available on line via the Orientations of Proteins in Membranes (OPM) database (also see supplemental Methods). Anisotropic Network Modeling (ANM) software (available on line) using default parameters was used to carry out predictions of molecular motions within the dimer and hexamer structures. The ANM is a type of elastic network model in which the protein is described by a coarse-grained representation of each residue as a bead connected to other residues based on a cutoff distance via a harmonic oscillator. The low frequency modes of vibration, i.e. the characteristic patterns or shapes in which the system will vibrate, obtained by the ANM correlate well with experimentally observed changes and have become a routine method of analysis of protein dynamics (40). Most systems have many modes of vibration, and the top modes predicted by the ANM have been inspected.
CL was docked to monomeric (chain A), dimeric and hexameric forms of Nm23-H4 (PDB code 1EHW) using Autodock Vina (41). Marvin Sketch 5.3.6 (2010, ChemAxon) was used for drawing, displaying the chemical structures and generating the three-dimensional structures corresponding to the lowest energy conformer. The AutoDockTools package was employed for pre-processing and converting the PDB files into pdbqt format. Pre-processing involved addition of hydrogens and charges to the protein molecule. The docking studies were performed using monomer, dimer, and hexamer conformations of Nm23-H4, using a grid box of size 42 × 46 × 40 Å, 60 × 66 × 60 Å, and 80 × 80 × 80 Å, respectively. The center of the molecule in each case was chosen as the grid center. The resulting predicted binding conformations were clustered together to estimate the total number of binding poses in each case. The probability of each cluster, lowest binding energy conformations, and electrostatic interaction between phosphate groups on CL and positively charged residues on the protein molecule for each pose were considered for further analysis.
RESULTS
Nm23-H4 Phosphotransfer Activity, Complexes with OPA1
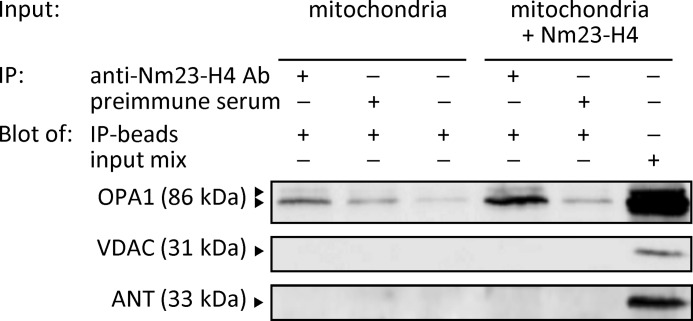
Nm23-H4 is the only isoform with NDPK activity localized inside mitochondria, but nucleotides generated by the cytosolic NDPK isoforms (Nm23-H1 to -H3) would in principle also have access to the IMS. We hypothesized that an advantage of Nm23-H4 phosphotransfer in the IMS could be constant regeneration of nucleoside triphosphates such as GTP driven by permanent ATP supply, and subsequent direct GTP delivery to mitochondrial GTPases. Nm23-H4/protein interactions were therefore analyzed by immunoprecipitation of complexes present in mitochondrial lysates (Fig. 2). Only the mitochondrial GTPase OPA1, but not the most abundant mitochondrial proteins VDAC or ANT (Fig. 2), were found complexed with Nm23-H4, thus supporting our hypothesis.
FIGURE 2.
Nm23-H4 interacts with OPA1. Nm24-H4 was immunoprecipitated (IP) from lysates of purified mitochondria or the same lysates complemented with recombinant Nm24-H4 by using isoform-specific anti-Nm24-H4 polyclonal antibodies (Ab) or corresponding preimmune serum. Immunoprecipitates were tested by immunoblotting for presence of OPA1, VDAC, and ANT. An input mix was tested as a positive control (right lane). Representative blots of three independent experiments are shown.
Nm23-H4 Phosphotransfer Activity, Inhibition by Anionic Lipids
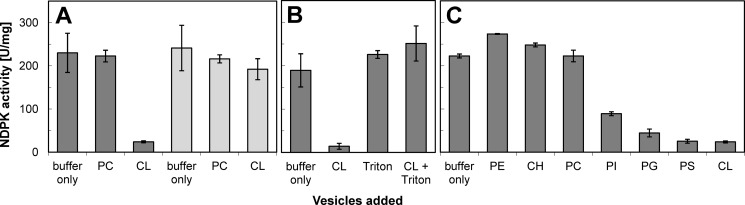
Next, we examined how Nm23-H4 phosphotransfer activity is affected by its membrane interactions. The effect of vesicles containing either anionic CL or zwitterionic phosphatidylcholine (PC) with neutral net charge was analyzed in an in vitro NDPK activity assay (Fig. 3A). Under conditions used in this assay, Nm23-H4 is quantitatively bound at both binding sides to the lipid vesicles. Surprisingly, CL inhibited Nm23-H4 to about 90%, although PC had no effect. Subsequent incubation with a detergent, Triton X-100, relieved CL inhibition of WT protein (Fig. 3B), showing that the protein/lipid interaction led to inhibition of phosphotransfer activity. The CL inhibition was not observed with the R90D mutant that does not interact with anionic phospholipids (8) or with cytosolic Nm23-H2, which also possesses an aspartate instead of a basic residue at the homologous position (data not shown). Because Nm23-H4 affinity with different anionic phospholipids is increasing in the order phosphatidylinositol < phosphatidylserine, phosphatidylglycerol, CL (KD ∼30 nm) (8), we analyzed the effect of these phospholipids and zwitterionic phospholipids with neutral net charge such as PC, phosphatidylethanolamine, or cholesterol (Fig. 3C). All anionic phospholipids showed a clear inhibitory effect that correlates with their binding to Nm23-H4 reported earlier (8).
FIGURE 3.
Nm23-H4 phosphotransfer activity is inhibited by anionic phospholipids. A, phosphotransfer activity of Nm23-H4 WT (dark gray bars) is inhibited to about 90% by binding of anionic CL (100 μg/ml), whereas zwitterionic PC (100 μg/ml) has no effect. By contrast, phosphotransfer activity of membrane binding-deficient Nm23-H4 R90D mutant (R90D, light gray bars) is not affected by CL. B, inhibition of Nm23-H4 WT phosphotransfer activity by CL can be relieved by addition of detergent (0.5% Triton X-100). C, phospholipids with neutral net charge, PC, and phosphatidylethanolamine (PE), or cholesterol (CH) do not inhibit Nm23-H4 phosphotransfer, although anionic phospholipids phosphatidylinositol (PI), phosphatidylglycerol (PG), phosphatidylserine (PS), and CL are inhibitory (all lipids at 100 μg/ml). Note: under conditions used in this assay, Nm23-H4 is quantitatively bound at both binding faces to lipid vesicles (8). Furthermore, this inhibition profile parallels the binding of Nm23-H4 to these lipids (see Fig. 2 in Ref. 8). Data are means ± S.D., n = 3.
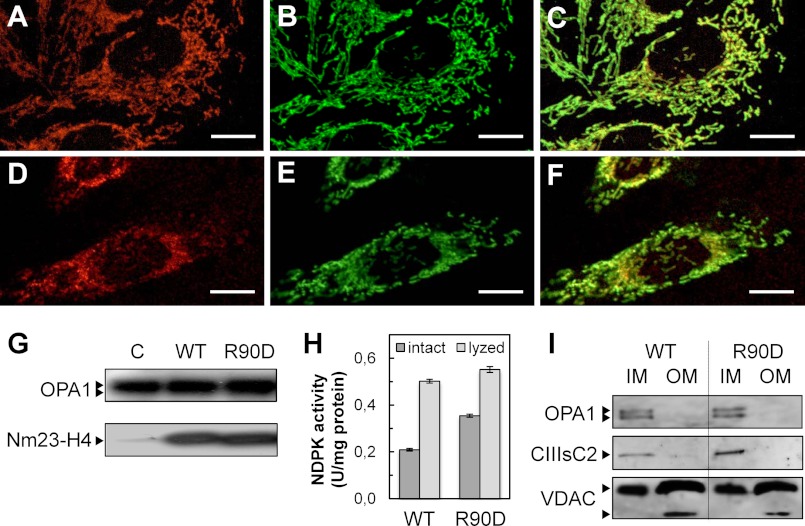
To analyze the relationship between phospholipid interaction and NDPK activity within mitochondria, we used an in vivo model system based on HeLa cells that express only faint amounts of immunodetectable endogenous Nm23-H4 (Fig. 4G). In these cells, stable expression of Nm23-H4 WT and the membrane binding-deficient R90D mutant led both proteins to correct mitochondrial localization (Fig. 4, A–F), similar expression level (Fig. 4G), and similar specific NDPK activity in detergent-lysed mitochondria (Fig. 4H). However, when specific NDPK activity was measured in isolated intact mitochondria, where substrates have largely free access to Nm23-H4 in the IMS, it was about 70% higher with R90D mitochondria as compared with WT controls (Fig. 4H). This suggests that mitochondrial phosphotransfer activity of Nm23-H4 WT was also inhibited in vivo relative to the R90D mutant, similar to what was observed with pure phospholipids in vitro (Fig. 3).
FIGURE 4.
Nm23-H4 WT and R90D mutant express at similar levels in mitochondria of HeLa cells. HeLa cells were stably transfected with vectors expressing Nm23-H4 wild-type (WT) or membrane binding-deficient mutant (R90D) after induction with tetracycline. A–F, confocal microscopy showing colocalization of mitochondrion-selective dye MitoTracker Red CMXRos (red) (A and D) with immunolabeled Nm23-H4 (green) (B and E) in HeLa clones expressing WT (A–C) or R90D (D–F). Colocalization of Nm23-H4 with mitochondria is shown in merged images (yellow) (C and F). Micrographs were obtained from a single focal section (scale bars, 10 μm). G, immunoblots for Nm23-H4 and mitochondrial OPA1 (loading control) in mitochondrial extracts of HeLa cells stably transfected with empty vector control (C) or vectors expressing WT or R90D. H, NDPK phosphotransfer activity in intact WT or R90D mitochondria and after mitochondrial lysis with 0.5% Triton X-100; data are means ± S.D., n = 3. I, immunoblots of mitochondrial IM and OM subfractions purified from HeLa cells expressing WT or R90D, probed for markers of IM (OPA1, Complex III subunit Core 2, CIIIsC2) and OM (VDAC; note: an additional short VDAC species co-purifies exclusively with OM (49)). Representative blots of three independent experiments are shown.
Evidence for in Vivo Nm23-H4 Lipid Transfer Activity, Altered CL Distribution
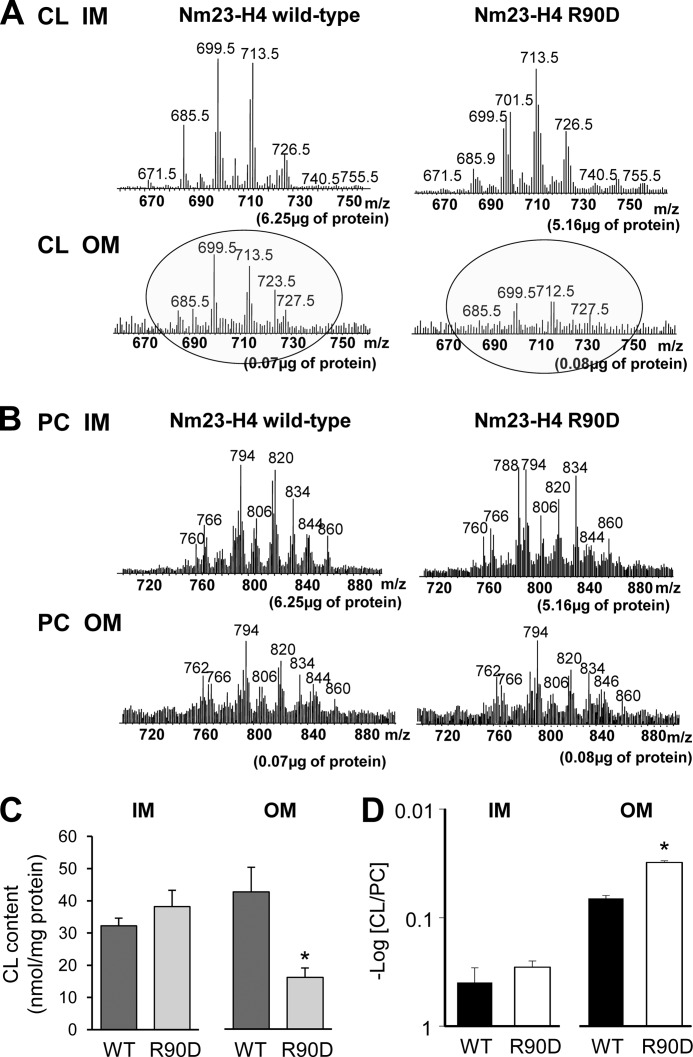
To explore in live cells the intermembrane lipid transfer activity of Nm23-H4 seen in vitro (6), we employed the same HeLa model system. Mitochondria from largely Nm23-H4-deficient HeLa cells expressing Nm23-H4 WT or R90D mutant were sub-fractioned into IM-enriched and pure OM fractions (Fig. 4I). HPLC-ESI-MS-based lipidomics were then applied to determine and quantify the PC and CL species in these membrane fractions (Tables 1 and 2 and Fig. 5). Although CL content was very low in pure OM of mitochondria harboring the R90D mutant, its level was clearly higher in cells expressing Nm23-H4 WT (Fig. 5A, circled spectra). Accordingly, a greater number of individual CL molecular species was detectable in the OM fraction from cells with WT Nm23-H4 than with the R90D mutant (Table 2). This was not based on the appearance of a new CL species but rather on redistribution of the existing species (Fig. 5A, see circled spectra). Quantification of MS data confirmed a significantly higher CL content only in the OM of Nm23-H4 WT mitochondria, irrespective of normalization to either protein (Fig. 5C) or PC content (Fig. 5D) of the corresponding membrane fractions. The redistribution of phospholipids between the IM and OM was CL-specific as the OM levels of the one most abundant phospholipid, PC, was similar in Nm23-H4 WT and R90D mitochondria (Fig. 5B). These data indicate that Nm23-H4 WT (but not R90D mutant) caused a decrease in the CL asymmetry between IM and OM.
TABLE 1.
Major molecular species of CL and PC in HeLa mitochondria expressing Nm23-H4 WT or R90D mutant
| m/z (M − 2H)2− | Molecular species |
|---|---|
| Cardiolipin | |
| 671.9541 | (16:1)4 |
| 684.4574 | (18:2)1(16:1)3 |
| 685.4651 | (18:1)1(16:1)3 |
| 686.4701 | (18:1)1(16:1)2(16:0)1 |
| 687.4794 | (18:1)1(16:1)1(16:0)2 |
| 698.4727 | (16:1)2(18:1)1(18:2)1 |
| 699.4803 | (16:1)2(18:1)2 |
| 700.4864 | (16:1)1(16:0)1(18:1)2 |
| 701.4985 | (16:1)1(16:0)1(18:1)1(18:0)1 |
| 710.4729 | (16:1)2(18:2)1(20:3)1 |
| 711.4811 | (16:1)1(18:2)2(18:1)1 |
| 712.4885 | (16:1)1(18:2)1(18:1)2 |
| 713.4959 | (16:1)1(18:1)3 |
| 714.5014 | (16:1)1(18:0)1(18:1)2 |
| 723.4802 | (18:2)4 |
| 724.4881 | (16:1)1(18:2)1(18:1)1(20:3)1 |
| 725.4964 | (18:2)2(18:1)2 |
| 726.5039 | (18:2)1(18:1)3 |
| 727.5104 | (18:1)4 |
| Phosphatidylcholine | |
| 766.5184 | (14:0)(18:1) |
| 794.5401 | (16:0)(18:1) |
| 820.5676 | (18:1)(18:1) |
| 834.6242 | (18:1)(22:4) |
| 844.5664 | (18:0)(20:4) |
| 860.6172 | (20:0)(22:6) |
TABLE 2.
Clusters in LC-MS spectra of CL from inner (IM) and outer membranes (OM) of HeLa mitochondria expressing Nm23-H4 WT or R90D mutant
| Nm23-H4 |
m/z cluster |
|||||||
|---|---|---|---|---|---|---|---|---|
| 671 | 685 | 691 | 699 | 713 | 723 | 726/727 740 | 755 | |
| WT IM | + | + | + | + | + | + | + + | + |
| WT OM | − | + | + | + | + | + | + − | − |
| R90D IM | + | + | + | + | + | + | + + | + |
| R90D OM | − | + | − | + | + | + | + − | − |
FIGURE 5.
Expression of Nm23-H4 wild-type (but not R90D mutant) in HeLa cells affects CL (but not PC) distribution in mitochondrial membranes. Electrospray ionization mass spectrometry (ESI-MS) of lipid species in mitochondrial IM and OM subfractions purified from HeLa cells stably expressing WT or R90D Nm23-H4. Representative ESI-MS spectra for CL (A) and PC (B). C, quantitative assessment of CL in IM and OM; values were normalized to protein in the corresponding fraction. D, CL/PC ratios in IM and OM; the ratios of CL (mol %) to PC (mol %) in samples of IM and OM were calculated. Note: a higher value of −log(CL/PC) corresponds to lower ratio values. Data are means ± S.D., n = 3; *, p < 0.02 (C) or p < 0.04 (D) versus WT.
Altered CL Distribution Correlates with Mitochondrial Bax Accumulation and Sensitivity for Apoptotic Triggers
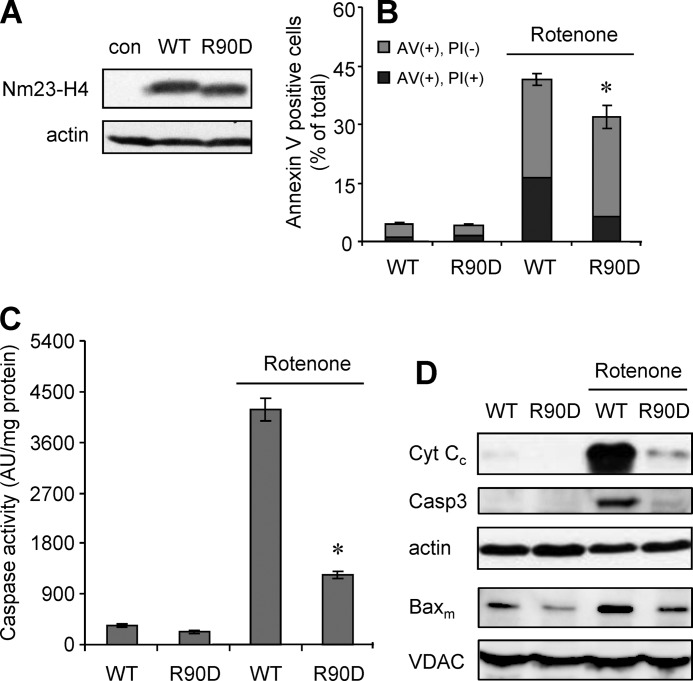
Redistribution of CL between the IM and OM and partial collapse of CL asymmetry is a requirement for the effective execution of the intrinsic apoptotic program (3, 21, 42). Therefore, we next examined mitochondrial Bax and apoptotic markers in HeLa cells stably expressing Nm23-H4 WT or R90D mutant (Fig. 6A). Rotenone treatment induced apoptosis in both cell types as determined by accumulation of mitochondrial Bax, cytochrome c release into the cytosol, caspase activation, cleaved caspase 3, and annexin V binding (Fig. 6, B–D), albeit at different levels. Expression of Nm23-H4 WT in HeLa cells, which increases OM CL content (Fig. 5B), led to much higher levels of mitochondrial Bax and significantly increased apoptosis as compared with R90D mutant. Mitochondria from WT cells revealed higher levels of Bax already under base-line conditions as compared with those from R90D cells (Fig. 6D). These data suggest that not only CL is transferred by Nm23-H4 to the OM but that it also participated in pro-apoptotic signaling.
FIGURE 6.
Expression of Nm23-H4 wild-type (but not R90D mutant) in HeLa cells increases apoptosis. Clones overexpressing comparable levels of Nm23-H4 WT or membrane binding-deficient mutant (R90D) without specific induction were used. After induction of apoptosis by addition of 1 μm rotenone, different apoptotic markers were determined. A, expression of Nm23-H4 proteins analyzed by immunoblot. B, cell staining for annexin V (AV, exposed phosphatidylserine) and propidium iodide (PI) as analyzed by FACS, identifying early apoptotic cells (annexin V-positive, propidium iodide-negative; gray bars) and dead cells (annexin V-positive, propidium iodide-positive; black bars). C, caspase 3/7 activity. D, cytochrome c released into the cytosol (Cyt Cc), cleaved caspase 3 (Casp3) and mitochondrial Bax (Baxm) were analyzed by immunoblot (actin and VDAC are cytosolic and mitochondrial loading controls, respectively). Data are means ± S.D., n = 3; *, p < 0.05 (B) or p < 0.01 (C) versus WT treated with rotenone.
Molecular Modeling of Nm23-H4 as a CL Transfer Nanomachine
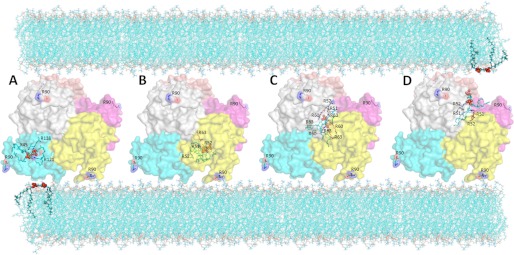
Our experimental data provide evidence for a dual function of Nm23-H4 in mitochondria. Although the mechanism of NDPK activity has been established decades ago (43), it is currently unknown how Nm23-H4 can facilitate intermembrane CL transfer. To gain mechanistic insight into the interactions of Nm23-H4 with the membrane, and with CL in particular, we used molecular modeling techniques. First, we predicted the orientation of Nm23-H4 with respect to the membrane using the OPM server (see “Experimental Procedures” and supplemental Fig. S1, A–C). As expected, Arg-90, in addition to several other residues nearby (Fig. 7 and supplemental Fig. S1, B and C), makes the primary contact with the membrane. Depicted in Fig. 7 is the predicted orientation with respect to the bottom membrane bilayer, although a top bilayer is indicated schematically. The orientation of the six Arg-90 residues of a hexamer suggests that either only a subset interacts with the IM and OM and/or that membrane curvature is required to facilitate interaction or that the protein orientation with respect to the membrane is dynamic. To test for the latter hypothesis, we carried out coarse grained dynamics calculations, using the Anisotropic Network Model (see Experimental Procedures). Mode 1 (see “Experimental Procedures”) is shown in supplemental Fig. S1D (a movie of the predicted motions is available as supplemental Movie). The most flexible region is the loop containing Arg-90. The predicted motions are such that, given the orientation with respect to the membrane predicted by OPM (Fig. 7), a rotation of the hexamer mediated by the paddling action of this loop would result in sequential interaction of this loop with the membranes.
FIGURE 7.
Molecular modeling of cardiolipin-binding sites in Nm23-H4 and putative lipid transfer mechanisms. The orientation of the hexamer is shown as predicted by OPM (also see supplemental Fig. S1). Different chains are indicated by different colors. Surface representation is used. Arg-90 is highlighted in all structures. CL is indicated in green, with phosphates highlighted as red spheres. In each model, arginine and lysine residues within 5 Å of the phosphates on CL are shown. A, most highly ranked and most frequently observed model in the docking of CL to either the monomer or the dimer structure of Nm23-H4. The bound conformation (model 1, ranked highest) is projected onto the hexamer structure here for ease of comparison of the binding pocket. B, binding pocket observed in 2/9 top ranked conformations of docking CL to the hexamer. C, most frequently observed and most highly ranked pocket in the hexamer. D, binding pocket observed in 2/9 top ranked conformation of docking CL to the hexamer, involving different chains than the conformation shown in B. It is related to the pocket shown in B by the symmetry of the hexamer, and there is significant overlap between the two pockets (see supplemental Table S1)
To gain insight into the mechanism by which Nm23-H4 could transfer CL from one bilayer to the other, we carried out molecular docking simulations. Here, CL is treated as an isolated molecule, not as part of the membrane, to identify specific pockets for CL within the Nm23-H4 structure. Using theoretical monomer and dimer as well as native hexamer models as the receptors for CL binding, several binding pockets were predicted (Fig. 7; all residues within 5 Å of the CL phosphate groups are listed in supplemental Table S1). In the monomer and dimer, the primary binding pocket involves interactions of the CL phosphate groups with Lys-45, Arg-121, and Arg-138 and extensive contacts with nearby residues, including the catalytic His-151 (Fig. 7A, shown projected onto the hexamer structure). The validity of this predicted site is supported by its overlap with the catalytic site; our experiments show that CL binding inhibits catalytic activity (Fig. 3). This pocket was not observed in the hexamer (Fig. 7, B–D). Instead, CL docked with highest rank in the center of the hexamer (Fig. 7C), making contacts with several copies of the same arginine residues contributed by different chains, in particular Arg-52, Arg-60, and Arg-63. There were also two additional sites, sandwiched between four chains in the crevice formed by the trimeric axes (Fig. 7, B and D). Because of the symmetry, these two sites are composed of highly similar residues, although one site shows additional overlap with the site observed in the central pocket (e.g. Arg-63, supplemental Table S1). These results suggest a model in which the high positive charge densities created by the hexamer interfaces facilitate transmission of CL through the hexamer, largely buried from the solvent (supplemental Fig. S2). It is tempting to speculate that the different pockets represent different stages in the pathway of CL going from one side of the membrane to the other (Fig. 7).
DISCUSSION
Currently, mitochondria are viewed not only as energy- and metabolite-generating organelles but also as complex integrators of cell signaling and death. Different mitochondrial proteins with diversified activities have evolved multiple activities to serve these various functions (2). Here, we provide evidence that Nm23-H4, the mitochondrial isoform of the Nm23 family of proteins, also belongs to this type of enzyme. Besides its energetic function in local NTP supply, Nm23-H4 participates in CL transfer from IM to OM with functions in apoptotic signaling.
Nm23-H4 is known for its NDP/NTP phosphotransfer activity. In contrast to the three cytosolic NDPK isoforms, mitochondrial Nm23-H4 phosphotransfer can be directly driven by mitochondrial ATP supply and deliver NTPs locally. Furthermore, complex formation of Nm23-H4 with the mitochondrial GTPase OPA1 reported here would allow GTP regeneration in direct vicinity of the GTPase. Similar cases of substrate channeling are well known for other mitochondrial kinases (44, 45). OPA1 is known to be essential for regulation of mitochondrial dynamics and function, including apoptosis (13, 14). Notably, the GTPase activity of OPA1 is strongly activated by anionic phospholipids, particularly CL (46). Recently, a new partner of OPA1-regulated mechanisms involved either in mitochondrial dynamics (fusion/fission) or in apoptosis, Bnip3, a mitochondrial proapoptotic BH3-only protein of the Bcl2 family, has been identified (13, 14).
Our data on binding of OPA1 with Nm23-H4 and possible regulation of both proteins by CL species add a new layer of complexity to this machinery as emphasized by two further findings of this study. First, we show that phosphotransfer activity of Nm23-H4 is strongly inhibited by binding to anionic phospholipids or mitochondrial membranes, an interaction that occurs in vivo preferentially at the IM (8), but it can also lead to membrane cross-linking (6, 8). Most probably, the proximity between the primary lipid-binding site and the phosphotransfer site (Fig. 1) leads to steric hindrance of nucleotide exchange upon membrane interaction. Second, we show that binding to the anionic phospholipid CL is necessary for Nm23-H4 to facilitate CL transfer in HeLa cells from its site of synthesis, the IM, to the OM and that this transfer enhances apoptotic cell death. This extends our earlier observations on in vitro intermembrane transfer of model lipids (6) to mitochondrial membranes in live cells.
The molecular modeling studies propose a putative mechanism for such CL transfer that can be tested. The predicted orientation of Nm23-H4 with respect to the membrane confirms the experimentally observed interaction with Arg-90 in the basic RRK triad. CL docking results, however, indicate that this may be only the initial CL interaction site in a series of CL interactions with additional sites within the Nm23-H4 structure. These may help to extract CL from its membrane environment. It is further conceivable that Nm23-H4 hexamers, existing in equilibrium of membrane- bound and, to a lesser extent, unbound states, can rebind after having rotated. This rotation could be mediated by a paddling motion predicted by the Anisotropic Network Model as the major dynamic property of Nm23-H4. The hereby conferred flexibility may expose bound CL to the opposite membrane for reintegration. Note, however, that there are several sites of interaction with CL predicted within the hexamer that are partially overlapping and that could thus be occupied in sequence. In this model, rotation is not a requirement of the CL transfer mechanism. Future studies are required to distinguish between these hypotheses, but the molecular modeling provides strong support for a structural basis enabling CL transfer provided by the hexameric assembly.
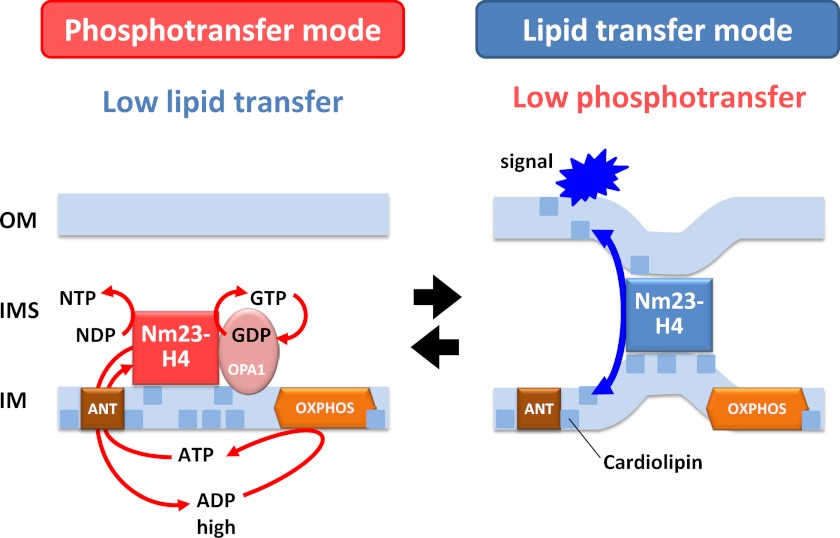
These and previously published data support a model where hexameric Nm23-H4 complexes are working as a lipid-dependent molecular nanoswitch (Fig. 8), either in a phosphotransfer or a lipid transfer mode. In phosphotransfer mode (Fig. 8, left panel), the enzyme is bound to the IM only as shown earlier (8). Such binding has a minor inhibitory effect on NDPK activity, because it occurs only at one binding face of the Nm23-H4 hexamer. By contrast, as we have shown, this topology favors ADP/ATP exchange via the IM adenylate translocase for stimulation of respiration (8), and it also directly provides GTP for mitochondrial GTPases like OPA1 (this study). However, it is insufficient to allow intermembrane CL transfer. In lipid transfer mode (Fig. 8, right panel), Nm23-H4 is fully bound to both membranes, i.e. at both binding faces as observed in vitro (6). Under these conditions, corresponding to those in our in vitro NDPK assay, phosphotransfer activity is strongly inhibited, whereas intermembrane CL transfer would be favored. As suggested by our data, CL occurring at the OM could increase accumulation of mitochondrial Bax and thus sensitize the cells for apoptotic stimuli. How exactly the switch between the two states and modes of action of Nm23-H4 works and what is the role of CLs in regulating Nm23-H4/OPA1 interactions will require further research. The additional binding to OM may depend on the availability or accessibility of CL and/or other anionic phospholipids in the IM outer leaflet and the OM inner leaflet.
FIGURE 8.
Model of Nm23-H4 as a bifunctional nanoswitch. In phosphotransfer mode (elements in red/brown/orange), Nm23-H4 only binds to mitochondrial IM and thus maintains phosphotransfer activity, although lipid transfer activity is low or absent. In this mode, Nm23-H4 regenerates NTP (mainly GTP) in the IMS, in particular for local use, e.g. by the interacting GTPase OPA1. Nm23-H4 also channels ADP via ANT into the matrix space for stimulation of respiration and ATP regeneration (8). In the lipid transfer mode (elements in light and dark blue), the symmetrical Nm23-H4 hexamers have to be fully membrane-bound, i.e. attached simultaneously to IM and OM, virtually cross-linking these two membranes (6, 8). This will inhibit phosphotransfer activity but allow for intermembrane lipid transfer, in particular of cardiolipin, whose exposure at the mitochondrial surface is suggested to have signaling character. The switch between the two modes may depend on the availability/accessibility of cardiolipin and other anionic phospholipids in the IM outer leaflet and the OM inner leaflet (for further details see text). OXPHOS, oxidative phosphorylation.
We propose that Nm23-H4 facilitates the partial collapse of CL asymmetry resulting in CL externalization on the mitochondrial surface. This can serve as a signal, similarly to externalization of phosphatidylserine resulting in the phagocytosis of apoptotic cells. The molecular mechanisms of kinase-driven CL transfer and its interactions with other CL transfer machineries like scramblase 3 (31), tBid (26), or Bax (33), as well as the signaling role of externalized CL and its oxidative modifications in mitophagy and apoptosis (24, 47) are attractive areas for further exploration.
This work was supported, in whole or in part, by National Institutes of Health Grants U19AIO68021, HL70755, NS076511, ES020693, and ES021068 (to V. K.) and Grant R01 LM007994-07 (to J. K. S.). This work was also supported by the French “Agence Nationale de Recherche” “Chaire d'Excellence” (to U. S.), National Institute for Occupational Safety and Health Grant OH008282 (to V. K.), National Science Foundation Grant CCF-1144281 (to J. K. S.), a grant from the “Groupement des Entreprises Françaises Contre le Cancer” (to M. L. L. and M. B.), and in part by the Canadian Natural Sciences and Engineering Research Council Grant 9848 (to R. M. E.).
This article was selected as a Paper of the Week.

This article contains supplemental Figs. S1 and S2, Movie, Table S1, and additional references.
- CL
- cardiolipin
- IM
- inner mitochondrial membrane
- IMS
- mitochondrial intermembrane space
- NDPK
- nucleoside diphosphate kinase
- Nm23
- nonmetastatic protein 23
- PC
- phosphatidylcholine
- OM
- outer mitochondrial membrane
- OPA1
- Optic Atrophy 1
- PDB
- Protein Data Bank
- Ap5A
- P1,P5-di(adenosine 5′)-pentaphosphate
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid
- ANM
- Anisotropic Network Modeling
- IP
- immunoprecipitation
- ANT
- adenylate translocase
- VDAC
- voltage-dependent anion channel.
REFERENCES
- 1. Nunnari J., Suomalainen A. (2012) Mitochondria. In sickness and in health. Cell 148, 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Galluzzi L., Kepp O., Trojel-Hansen C., Kroemer G. (2012) Non-apoptotic functions of apoptosis-regulatory proteins. EMBO Rep. 13, 322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kagan V. E., Tyurin V. A., Jiang J., Tyurina Y. Y., Ritov V. B., Amoscato A. A., Osipov A. N., Belikova N. A., Kapralov A. A., Kini V., Vlasova I. I., Zhao Q., Zou M., Di P., Svistunenko D. A., Kurnikov I. V., Borisenko G. G. (2005) Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 1, 223–232 [DOI] [PubMed] [Google Scholar]
- 4. Hüttemann M., Pecina P., Rainbolt M., Sanderson T. H., Kagan V. E., Samavati L., Doan J. W., Lee I. (2011) The multiple functions of cytochrome c and their regulation in life and death decisions of the mammalian cell. From respiration to apoptosis. Mitochondrion 11, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Milon L., Meyer P., Chiadmi M., Munier A., Johansson M., Karlsson A., Lascu I., Capeau J., Janin J., Lacombe M. L. (2000) The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase. J. Biol. Chem. 275, 14264–14272 [DOI] [PubMed] [Google Scholar]
- 6. Epand R. F., Schlattner U., Wallimann T., Lacombe M. L., Epand R. M. (2007) Novel lipid transfer property of two mitochondrial proteins that bridge the inner and outer membranes. Biophys. J. 92, 126–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Milon L., Rousseau-Merck M. F., Munier A., Erent M., Lascu I., Capeau J., Lacombe M. L. (1997) nm23-H4, a new member of the family of human nm23/nucleoside diphosphate kinase genes localized on chromosome 16p13. Hum. Genet. 99, 550–557 [DOI] [PubMed] [Google Scholar]
- 8. Tokarska-Schlattner M., Boissan M., Munier A., Borot C., Mailleau C., Speer O., Schlattner U., Lacombe M. L. (2008) The nucleoside diphosphate kinase D (NM23-H4) binds the inner mitochondrial membrane with high affinity to cardiolipin and couples nucleotide transfer with respiration. J. Biol. Chem. 283, 26198–26207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lacombe M. L., Tokarska-Schlattner M., Epand R. F., Boissan M., Epand R. M., Schlattner U. (2009) Interaction of NDPK-D with cardiolipin-containing membranes. Structural basis and implications for mitochondrial physiology. Biochimie 91, 779–783 [DOI] [PubMed] [Google Scholar]
- 10. Schlattner U., Tokarska-Schlattner M., Ramirez S., Brückner A., Kay L., Polge C., Epand R. F., Lee R. M., Lacombe M. L., Epand R. M. (2009) Mitochondrial kinases and their molecular interaction with cardiolipin. Biochim. Biophys. Acta 1788, 2032–2047 [DOI] [PubMed] [Google Scholar]
- 11. Lacombe M. L., Milon L., Munier A., Mehus J. G., Lambeth D. O. (2000) The human Nm23/nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 32, 247–258 [DOI] [PubMed] [Google Scholar]
- 12. Boissan M., Dabernat S., Peuchant E., Schlattner U., Lascu I., Lacombe M. L. (2009) The mammalian Nm23/NDPK family. From metastasis control to cilia movement. Mol. Cell. Biochem. 329, 51–62 [DOI] [PubMed] [Google Scholar]
- 13. Landes T., Emorine L. J., Courilleau D., Rojo M., Belenguer P., Arnauné-Pelloquin L. (2010) The BH3-only Bnip3 binds to the dynamin Opa1 to promote mitochondrial fragmentation and apoptosis by distinct mechanisms. EMBO Rep. 11, 459–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fang H. Y., Chen C. Y., Chiou S. H., Wang Y. T., Lin T. Y., Chang H. W., Chiang I. P., Lan K. J., Chow K. C. (2012) Overexpression of optic atrophy 1 protein increases cisplatin resistance via inactivation of caspase-dependent apoptosis in lung adenocarcinoma cells. Hum. Pathol. 43, 105–114 [DOI] [PubMed] [Google Scholar]
- 15. Ardail D., Privat J. P., Egret-Charlier M., Levrat C., Lerme F., Louisot P. (1990) Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 265, 18797–18802 [PubMed] [Google Scholar]
- 16. Claypool S. M., Koehler C. M. (2012) The complexity of cardiolipin in health and disease. Trends Biochem. Sci. 37, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Osman C., Voelker D. R., Langer T. (2011) Making heads or tails of phospholipids in mitochondria. J. Cell. Biol. 192, 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuwana T., Mackey M. R., Perkins G., Ellisman M. H., Latterich M., Schneiter R., Green D. R., Newmeyer D. D. (2002) Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111, 331–342 [DOI] [PubMed] [Google Scholar]
- 19. Gonzalvez F., Pariselli F., Dupaigne P., Budihardjo I., Lutter M., Antonsson B., Diolez P., Manon S., Martinou J. C., Goubern M., Wang X., Bernard S., Petit P. X. (2005) tBid interaction with cardiolipin primarily orchestrates mitochondrial dysfunctions and subsequently activates Bax and Bak. Cell Death Differ. 12, 614–626 [DOI] [PubMed] [Google Scholar]
- 20. Gonzalvez F., Schug Z. T., Houtkooper R. H., MacKenzie E. D., Brooks D. G., Wanders R. J., Petit P. X., Vaz F. M., Gottlieb E. (2008) Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J. Cell. Biol. 183, 681–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sorice M., Manganelli V., Matarrese P., Tinari A., Misasi R., Malorni W., Garofalo T. (2009) Cardiolipin-enriched raft-like microdomains are essential activating platforms for apoptotic signals on mitochondria. FEBS Lett. 583, 2447–2450 [DOI] [PubMed] [Google Scholar]
- 22. Landeta O., Landajuela A., Gil D., Taneva S., Di Primo C., Sot B., Valle M., Frolov V. A., Basañez G. (2011) Reconstitution of proapoptotic BAK function in liposomes reveals a dual role for mitochondrial lipids in the BAK-driven membrane permeabilization process. J. Biol. Chem. 286, 8213–8230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kagan V. E., Tyurina Y. Y., Bayir H., Chu C. T., Kapralov A. A., Vlasova I. I., Belikova N. A., Tyurin V. A., Amoscato A., Epperly M., Greenberger J., Dekosky S., Shvedova A. A., Jiang J. (2006) The “pro-apoptotic genies” get out of mitochondria: oxidative lipidomics and redox activity of cytochrome c/cardiolipin complexes. Chem. Biol. Interact. 163, 15–28 [DOI] [PubMed] [Google Scholar]
- 24. Kagan V. E., Bayir H. A., Belikova N. A., Kapralov O., Tyurina Y. Y., Tyurin V. A., Jiang J., Stoyanovsky D. A., Wipf P., Kochanek P. M., Greenberger J. S., Pitt B., Shvedova A. A., Borisenko G. (2009) Cytochrome c/cardiolipin relations in mitochondria. A kiss of death. Free Radic. Biol. Med. 46, 1439–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kapralov A. A., Yanamala N., Tyurina Y. Y., Castro L., Samhan-Arias A., Vladimirov Y. A., Maeda A., Weitz A. A., Peterson J., Mylnikov D., Demicheli V., Tortora V., Klein-Seetharaman J., Radi R., Kagan V. E. (2011) Topography of tyrosine residues and their involvement in peroxidation of polyunsaturated cardiolipin in cytochrome c/cardiolipin peroxidase complexes. Biochim. Biophys. Acta 1808, 2147–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Esposti M. D., Erler J. T., Hickman J. A., Dive C. (2001) Bid, a widely expressed proapoptotic protein of the Bcl-2 family, displays lipid transfer activity. Mol. Cell. Biol. 21, 7268–7276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gilquin B., Taillebourg E., Cherradi N., Hubstenberger A., Gay O., Merle N., Assard N., Fauvarque M. O., Tomohiro S., Kuge O., Baudier J. (2010) The AAA+ ATPase ATAD3A controls mitochondrial dynamics at the interface of the inner and outer membranes. Mol. Cell. Biol. 30, 1984–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li S., Rousseau D. (2012) ATAD3, a vital membrane-bound mitochondrial ATPase involved in tumor progression. J. Bioenerg. Biomembr. 44, 189–197 [DOI] [PubMed] [Google Scholar]
- 29. Tamura Y., Onguka O., Hobbs A. E., Jensen R. E., Iijima M., Claypool S. M., Sesaki H. (2012) Role for two conserved intermembrane space proteins, Ups1p and Up2p, in intra-mitochondrial phospholipid trafficking. J. Biol. Chem. 287, 15205–15218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Connerth M., Tatsuta T., Haag M., Klecker T., Westermann B., Langer T. (2012) Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science 338, 815–818 [DOI] [PubMed] [Google Scholar]
- 31. Liu J., Dai Q., Chen J., Durrant D., Freeman A., Liu T., Grossman D., Lee R. M. (2003) Phospholipid scramblase 3 controls mitochondrial structure, function, and apoptotic response. Mol. Cancer Res. 1, 892–902 [PubMed] [Google Scholar]
- 32. Bevers E. M., Williamson P. L. (2010) Phospholipid scramblase. An update. FEBS Lett. 584, 2724–2730 [DOI] [PubMed] [Google Scholar]
- 33. Epand R. F., Martinou J. C., Montessuit S., Epand R. M. (2003) Transbilayer lipid diffusion promoted by Bax. Implications for apoptosis. Biochemistry 42, 14576–14582 [DOI] [PubMed] [Google Scholar]
- 34. Tokarska-Schlattner M., Dolder M., Gerber I., Speer O., Wallimann T., Schlattner U. (2007) Reduced creatine-stimulated respiration in doxorubicin challenged mitochondria. Particular sensitivity of the heart. Biochim. Biophys. Acta 1767, 1276–1284 [DOI] [PubMed] [Google Scholar]
- 35. Agarwal R. P., Robison B., Parks R. E., Jr. (1978) Nucleoside diphosphokinase from human erythrocytes. Methods Enzymol. 51, 376–386 [DOI] [PubMed] [Google Scholar]
- 36. Hovius R., Lambrechts H., Nicolay K., de Kruijff B. (1990) Improved methods to isolate and subfractionate rat liver mitochondria. Lipid composition of the inner and outer membrane. Biochim. Biophys. Acta. 1021, 217–226 [DOI] [PubMed] [Google Scholar]
- 37. Walzel B., Speer O., Zanolla E., Eriksson O., Bernardi P., Wallimann T. (2002) Novel mitochondrial creatine transport activity. Implications for intracellular creatine compartments and bioenergetics. J. Biol. Chem. 277, 37503–37511 [DOI] [PubMed] [Google Scholar]
- 38. Folch J., Lees M., Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 39. Tyurina Y. Y., Kisin E. R., Murray A., Tyurin V. A., Kapralova V. I., Sparvero L. J., Amoscato A. A., Samhan-Arias A. K., Swedin L., Lahesmaa R., Fadeel B., Shvedova A. A., Kagan V. E. (2011) Global phospholipidomics analysis reveals selective pulmonary peroxidation profiles upon inhalation of single-walled carbon nanotubes. ACS Nano 5, 7342–7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Isin B., Tirupula K. C., Oltvai Z. N., Klein-Seetharaman J., Bahar I. (2012) Identification of motions in membrane proteins by elastic network models and their experimental validation. Methods Mol. Biol. 914, 285–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Trott O., Olson A. J. (2010) AutoDock Vina. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sorice M., Circella A., Cristea I. M., Garofalo T., Di Renzo L., Alessandri C., Valesini G., Esposti M. D. (2004) Cardiolipin and its metabolites move from mitochondria to other cellular membranes during death receptor-mediated apoptosis. Cell Death Differ. 11, 1133–1145 [DOI] [PubMed] [Google Scholar]
- 43. Garces E., Cleland W. W. (1969) Kinetic studies of yeast nucleoside diphosphate kinase. Biochemistry 8, 633–640 [DOI] [PubMed] [Google Scholar]
- 44. Schlattner U., Tokarska-Schlattner M., Wallimann T. (2006) Mitochondrial creatine kinase in human health and disease. Biochim. Biophys. Acta 1762, 164–180 [DOI] [PubMed] [Google Scholar]
- 45. Stachowiak O., Schlattner U., Dolder M., Wallimann T. (1998) Oligomeric state and membrane binding behaviour of creatine kinase isoenzymes: implications for cellular function and mitochondrial structure. Mol. Cell. Biochem. 184, 141–151 [PubMed] [Google Scholar]
- 46. Ban T., Heymann J. A., Song Z., Hinshaw J. E., Chan D. C. (2010) OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum. Mol. Genet. 19, 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bayir H., Fadeel B., Palladino M. J., Witasp E., Kurnikov I. V., Tyurina Y. Y., Tyurin V. A., Amoscato A. A., Jiang J., Kochanek P. M., DeKosky S. T., Greenberger J. S., Shvedova A. A., Kagan V. E. (2006) Apoptotic interactions of cytochrome c. Redox flirting with anionic phospholipids within and outside of mitochondria. Biochim Biophys. Acta 1757, 648–659 [DOI] [PubMed] [Google Scholar]
- 48. Dumas C., Lascu I., Moréra S., Glaser P., Fourme R., Wallet V., Lacombe M. L., Véron M., Janin J. (1992) X-ray structure of nucleoside diphosphate kinase. EMBO J. 11, 3203–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brahimi-Horn M. C., Ben-Hail D., Ilie M., Gounon P., Rouleau M., Hofman V., Doyen J., Mari B., Shoshan-Barmatz V., Hofman P., Pouysségur J., Mazure N. M. (2012) Expression of a truncated active form of VDAC1 in lung cancer associates with hypoxic cell survival and correlates with progression to chemotherapy resistance. Cancer Res. 72, 2140–2150 [DOI] [PubMed] [Google Scholar]