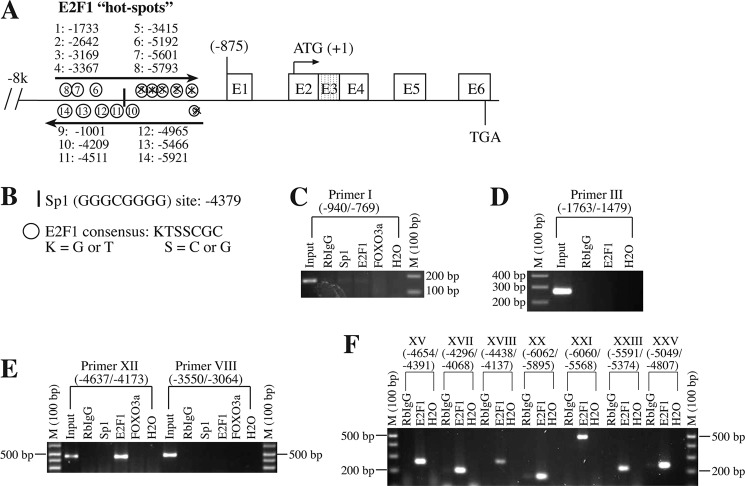
FIGURE 3.
E2F1 binds to the Bim promoter at multiple sites. A and B, putative E2F1-binding sites on the Bim promoter were determined by in silico analysis using KTSSCGC consensus sequence. C–F, ChIP analysis determines multiple E2F1-binding sites on the Bim promoter. Briefly, cancer cells were subjected to chromatin cross-linking followed by immunoprecipitation with anti-E2F1, anti-Sp1, anti-FOXO3a, and RbIgG. Bim promoter fragments were amplified by using PCR primers as described in Table 1. Positive controls (input DNA) or negative controls (H2O) were used in the amplification of the Bim promoter. Crossed sites in A (i.e. 1–5 and 9 predicted sites) indicate that these sites do not show binding with E2F1 based on ChIP analysis.