Background: The mechanism of cataract formation by the recently discovered γD-crystallin W42R mutant is unknown.
Results: Structural, biochemical, and biophysical studies revealed a partially unfolded species of the W42R mutant.
Conclusion: Partially unfolded species serve as nuclei for aggregation.
Significance: The properties of the W42R mutant γD-crystallin provide the link to the pathogenesis of age-related cataract caused by photodamaged wild-type γD-crystallin.
Keywords: Cataract, Crystallin, Mutant, NMR, X-ray Crystallography, Trypsin Digestion, UV Irradiation
Abstract
Some mutants of human γD-crystallin are closely linked to congenital cataracts, although the detailed molecular mechanisms of mutant-associated cataract formation are generally not known. Here we report on a recently discovered γD-crystallin mutant (W42R) that has been linked to autosomal dominant, congenital cataracts in a Chinese family. The mutant protein is much less soluble and stable than wild-type γD-crystallin. We solved the crystal structure of W42R at 1.7 Å resolution, which revealed only minor differences from the wild-type structure. Interestingly, the W42R variant is highly susceptible to protease digestion, suggesting the presence of a small population of partially unfolded protein. This partially unfolded species was confirmed and quantified by NMR spectroscopy. Hydrogen/deuterium exchange experiments revealed chemical exchange between the folded and unfolded species. Exposure of wild-type γD-crystallin to UV caused damage to the N-terminal domain of the protein, resulting in very similar proteolytic susceptibility as observed for the W42R mutant. Altogether, our combined data allowed us to propose a model for W42R pathogenesis, with the W42R mutant serving as a mimic for photodamaged γD-crystallin involved in age-related cataract.
Introduction
Cataract, referred to as opacification of all or part of the crystalline lens of the eye (1), can be broadly divided into two classes: early onset (congenital or juvenile) and age-related cataract (2). The congenital cataracts occur at an estimated prevalence of 2.2–2.5 cases per 10,000 live births (3), and ∼50% of congenital cataracts are inherited (4). Cataract-associated mutations in the crystallin genes are known to occur in all three crystallin families (5, 6) and show a variety of phenotypes. At the present time, at least 60 cases of inherited cataracts have been documented; these appear to be caused by a single-gene disorder, and amino acid substitutions have been mapped to the affected genes (7).
Human γD-crystallin (HGD)2 is the second most abundant γ-crystallin of the lens nucleus (8). It is monomeric and contains 173 amino acids. The overall architecture of HGD shows two highly homologous, duplicated β-sheet domains, each with two buried tryptophans: Trp-42 and Trp-68 in the N-terminal domain (N-td) and Trp-130 and Trp-156 in the C-terminal domain (C-td). These four tryptophans are conserved in all vertebrate γ-crystallins.
Mutations of hydrophobic residues associated with congenital or early onset cataract in mice have been identified, including F9S in murine γS-crystallin (9–11) and V75D (12) and I90F (13) in murine γD-crystallin. King and co-workers (14) performed systematic mutational studies on HGD, revealing the critical role of the hydrophobic core and important features of the domain interface for folding and stability of HGD. These mutants, including F56A, I81A, V132A, and L145A, possess a destabilized N-td that results in a non-negligible population of partially folded species. Indeed, it was observed that under mildly denaturing conditions, one domain can be folded and the other domain unfolded (14–17).
For other crystallin mutants such as murine γS-F9S (9) and human γC-T5P (18), a transient unfolding intermediate was observed under physiological conditions (19). R2 relaxation dispersion experiments on the murine γS-F9S mutant showed an ∼2% population of a loosely structured intermediate at 37 °C that increased with temperature (20). However, no such data exist for any human γD-crystallin variant.
Human eyes are directly exposed to ambient sunlight, including potentially damaging UV. In native crystallins, which constitute the major eye lens proteins, it is assumed that extensive tryptophan fluorescence quenching influences the lens response to ultraviolet light and protects the retina from ambient ultraviolet damage (21–23). Indeed, oxidized tryptophan has been identified in HGD in the mature-onset cataractous human lens by mass spectrometry (24), and tryptophan photodamage in the crystallins may be one of the factors in the etiology of cataract formation (25).
Recently, a novel mutation in HGD was discovered in a Chinese family, with the affected individuals exhibiting nuclear congenital cataracts (26). A single T/C change at position 127 in the CRYGD gene was mapped, translating into a tryptophan to arginine change at position 42 (W42R) in the amino acid sequence of the mature protein, i.e. the loss of one of the four conserved tryptophans from the protein. The W42R mutant protein was reported to be less stable and more prone to aggregate when subjected to environmental stresses, such as heat and UV irradiation, when compared with HGD (26). However, no structural or other biochemical characterizations were carried out to elucidate how this mutation could lead to cataract formation.
To address whether the loss of the tryptophan residue in the mutant alters the protein such as to make it more susceptible to aggregation and precipitation, we investigated W42R γD-crystallin by NMR spectroscopy and determined its high resolution x-ray structure. In addition, the thermodynamic behavior was probed by guanidinium hydrochloride (GdnHCl)-induced unfolding, and a comparative proteolysis study was performed for W42R and HGD. We show that W42R exhibits the HGD fold with no major, discernable conformational differences between mutant and wild type, although the relative domain orientation in the two x-ray structures is slightly different. Interestingly, however, a small, but noteworthy fraction of partially unfolded protein was detected for the W42R mutant in solution by NMR spectroscopy. Based on our overall results, we conclude that the W42R mutation introduces partial unfolding in the N-td, mimicking the effect of photodamage caused by UV exposure of HGD. This partially folded species may act as the seed for aggregation and precipitation of the mutant protein, nucleating cataract formation.
EXPERIMENTAL PROCEDURES
Expression and Purification of Proteins
Cloning, expression, and purification of the W42R mutant was essentially similar to that previously reported for wild-type HGD and the R76S mutant (27, 28). Briefly, Escherichia coli cells that harbored pET-14b with the mutant γD-crystallin gene were grown at 37 °C, induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside, and further grown at 16 °C for 16 h. Cells were then harvested by centrifugation and resuspended in 50 mm Tris buffer (pH 8.0), containing 1% (v/v) 100× HaltTM protease inhibitor mixture (Thermo Scientific), 1 mm DTT, 1 mm PMSF, and 1 mm EDTA, and lysed by passage through a Microfluidizer (Microfluidics, Newton, MA). The cell lysate was clarified by centrifugation at 100,000 × g for 1 h, and the supernatant was loaded onto a HiTrap Q XL anion exchange column (GE Healthcare) in 50 mm Tris buffer (pH 8.0), 1 mm EDTA, 1 mm DTT. The flow-through fraction was collected and dialyzed overnight against 10 mm MES buffer (pH 6.2) in the presence of 1 mm EDTA, 1 mm DTT, and 2% glycerol. The dialyzed sample was loaded on a HiTrap SP cation exchange column (GE Healthcare) and eluted using a linear NaCl gradient from 0 to 1 m over a 20-column volume. Fractions containing the protein were collected, concentrated, and subjected to gel filtration on a Superdex 75 26/60 column (GE Healthcare) in 20 mm sodium phosphate buffer (pH 6.2), 1 mm EDTA, 5 mm DTT as the final purification step.
All purified proteins were characterized by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry on a Voyager-DE PRO instrument, operated in a linear mode with external calibration. HGD and W42R exhibited experimental molecular masses of 20,592.10 and 20,558.05 Da, respectively (the theoretical molecular masses are 20,606.94 and 20,576.91 Da, respectively). Protein concentrations were determined using a NanoDrop 1000 spectrophotometer (Thermo Scientific), using extinction coefficients of 42.86 and 37.36 mm−1 cm−1 at 280 nm for HGD and W42R, respectively.
Crystallization
Small and low quality crystals of W42R were initially grown using the sitting drop vapor diffusion method from a solution containing 0.17 m Li2SO4, 0.085 m Tris-HCl (pH 8.5), 25.5% (w/v) PEG 4000, and 15% (v/v) glycerol. Subsequent optimization was performed by adding 1% (w/v) of octyl β-d-glucopyranoside to the protein solution (8, 12, and 16 mg/ml) and by varying both Li2SO4 (0.14, 0.18, 0.22, and 0.26 m) and PEG 4000 (18, 20, 22, 24, 26, and 28%) concentrations. The ratio of protein to mother liquor was kept constant at 2:2 μl. The final diffraction data were collected from a crystal grown in 0.18 m Li2SO4, 0.1 m Tris-HCl (pH 8.5), 22% (w/v) PEG 4000, and 15% (v/v) glycerol at a protein concentration of 16 mg/ml.
X-ray Data Collection and Processing
X-ray diffraction data were collected up to 1.6 Å on flash-cooled crystals (−180 °C) using a Rigaku FR-E generator with Saturn 944 charge-coupled device detectors at the wavelength corresponding to the copper edge (1.54 Å). Initial indexing of the diffraction patterns indicated that W42R crystallized in the P3221 space group with unit cell dimensions: a = 68.61 Å, b = 68.61 Å, and c = 133.38 Å (α = β = 90° and γ = 120°). Two molecules are present per asymmetric unit with an estimated solvent content of ∼44.11% (Vm = 2.2 Å3/Da) based on the Matthews Probability Calculator (29). All diffraction data were processed, integrated, and scaled using the d*TREK software (30) and eventually converted to MTZ format using the CCP4 package (31).
The structure for W42R was solved by molecular replacement using wild-type HGD (Protein Data Bank (PDB) ID: 1HK0) as the search model in PHASER (32). After generation of the initial model, the chain was rebuilt using Coot (33) followed by iterative refinement that involved alternating between manual rebuilding and automated refinement in REFMAC (34). Due to relatively incomplete reflections and intensity decay (high Rmerge) at the highest resolution range (1.7–1.6 Å), the final W42R structure was refined to 1.70 Å, with a final R-factor value of 20.0% and a free R value of 24.5%. 97.7 and 100% of all residues are located in the favored and allowed regions of the Ramachandran plot, respectively, and no residues were found in the disallowed region, as judged by MOLPROBITY (35). Pertinent data collection and refinement statistics are listed in Table 1. All structure figures were generated using the program PyMOL (36).
TABLE 1.
Data collection and refinement statistics (molecular replacement) for W42R
| Data collection | |
| Space group | P3221 |
| Cell dimensions | |
| a, b, c (Å) | 68.61, 68.61, 133.38 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 33.22–1.60 (1.66–1.60) |
| Rmerge | 0.115 (0.574) |
| 〈I/σI〉 | 11.2 (2.1) |
| Completeness (%) | 92.1 (84.0) |
| 〈Redundancy〉 | 10.59 (6.33) |
| Refinement | |
| Resolution (Å) | 30.50–1.70 (1.74–1.70) |
| No. of reflections | 36,152 (2495) |
| Rwork/Rfree | 0.200/0.245 (0.315/0.418) |
| No. of atoms | |
| Protein | 2932 |
| Water | 235 |
| B-factors | |
| Protein | 27.41 |
| Water | 35.20 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.081 |
Chemical Denaturation
Chemical unfolding by GdnHCl was monitored by tryptophan fluorescence, using an excitation wavelength of 295 nm, and emission was measured over 305–420 nm. Spectra were recorded at 37 °C on a Cary Eclipse fluorescence spectrophotometer (Agilent Technology Inc., Santa Clara, CA), equipped with a circulating water bath. Samples were prepared at 10 μg/ml protein concentration in 100 mm sodium phosphate buffer (pH 7.0), 5 mm DTT, 1 mm EDTA in the absence or presence of increasing GdnHCl concentrations (0–5.5 m). Prior to the measurements, the samples were incubated at 37 °C for 24 h to ensure that equilibrium had been reached. Spectra were corrected for background fluorescence from GdnHCl, and the unfolding curve was obtained by plotting the 360:320 nm fluorescence intensity ratio versus the concentration of GdnHCl. Equivalent data were collected for the urea unfolding experiments, conducted over the concentration range of 0–3 m urea.
Trypsin Digestion
HGD and W42R proteins (20 μg) were incubated overnight at 37 °C in the presence and absence of sequencing-grade trypsin (Promega, Madison, WI) at an enzyme-to-substrate ratio of 1:100, in 20 mm sodium phosphate buffer, pH 7.0, 5 mm DTT, 0.02% NaN3.
UV-C Irradiation
Solutions of unlabeled wild-type protein (1 mg/ml) in a 3-ml quartz cuvette were exposed to UV light of 254 nm at different total energy doses (1.0, 2.0, 3.0, 4.0, 5.0 J/cm2) using a SpectrolinkerTM UV source (Spectronics Corp.). Following irradiation, a small aliquot of each sample was saved, whereas the remainder was digested with trypsin as described above. Electrophoretic analysis was carried out for all irradiated samples, both with and without trypsin digestion. Similarly, a solution of 15N-labeled HGD (2 mg/ml) in an NMR tube was also exposed to UV light of 254 nm at an energy dose of 1.0 J/cm2. Like unlabeled HGD, a small aliquot was saved, and another aliquot was digested with trypsin. Both samples were analyzed on SDS-PAGE. For the remainder of the sample, a two-dimensional 1H-15N HSQC spectrum was recorded and compared with that of nonirradiated 15N-labeled HGD.
NMR Spectroscopy
NMR spectra for resonance assignments were acquired at 25 °C, using both 15N-labeled and 13C,15N-labeled samples on Bruker AVANCE 900, AVANCE 800, or AVANCE 600 spectrometers, equipped with 5-mm triple-resonance z-axis gradient cryoprobes. Temperature calibration was performed using the chemical shift differences between methyl and hydroxyl protons of 100% methanol. All samples contained 0.1 mm protein in NMR buffer (20 mm phosphate buffer (pH 6.2), 0.02% NaN3, 5 mm DTT, 5% (v/v) D2O). HNCA and HNCACB experiments were recorded and used for backbone resonance assignments. The combined amide proton and nitrogen chemical shift differences between mutant and HGD were calculated using Δδ = √((0.15 × ΔδN)2+(ΔδH)2). For hydrogen/deuterium (H/D) exchange experiments, 1H-15N HSQC spectra were recorded at 25 °C over a total time interval of 30 h on 15N-labeled 0.1 mm protein samples in NMR buffer in 50% (v/v) D2O at 600 MHz. Time points of 0, 6, 11, 22, and 30 h were chosen for analysis. Temperature-dependent 1H-15N HSQC spectra were recorded at 25, 37, and 42 °C at 900 MHz. All spectra were processed with NMRPipe (37) and analyzed using CARA (38) or SPARKY3 (version 3.113) (39).
RESULTS
Protein Purification and Solubility
Native recombinant W42R mutant protein without a His-tag was prepared because we previously observed that the presence of a His-tag significantly increased the solubility of the P23T γD-crystallin (27). Therefore, to assess the solubility and stability of the W42R mutant, we incubated the native mutant protein solution, concentrated at 10 mg/ml, and the wild-type protein as a control, at 37 °C overnight. After removal of protein precipitation by centrifugation, observed solely in the mutant sample, the final protein concentration in the supernatant was found to be 2.3 mg/ml. This is different from our findings for the wild-type protein, for which the solution stays clear and at the same protein concentration as initially measured. Indeed, the wild-type protein can be concentrated to ∼350 mg/ml, whereas the mutant protein readily precipitates when the protein concentration reaches ∼20 mg/ml. Our finding is therefore different from a previous report that found no noticeable difference between HGD and W42R solubility (26). Note that both protein constructs that were used in the previous study contained His tags and that the analysis was carried out at low protein concentration (5 mg/ml) at a single temperature (26).
Structure Assessment Using NMR
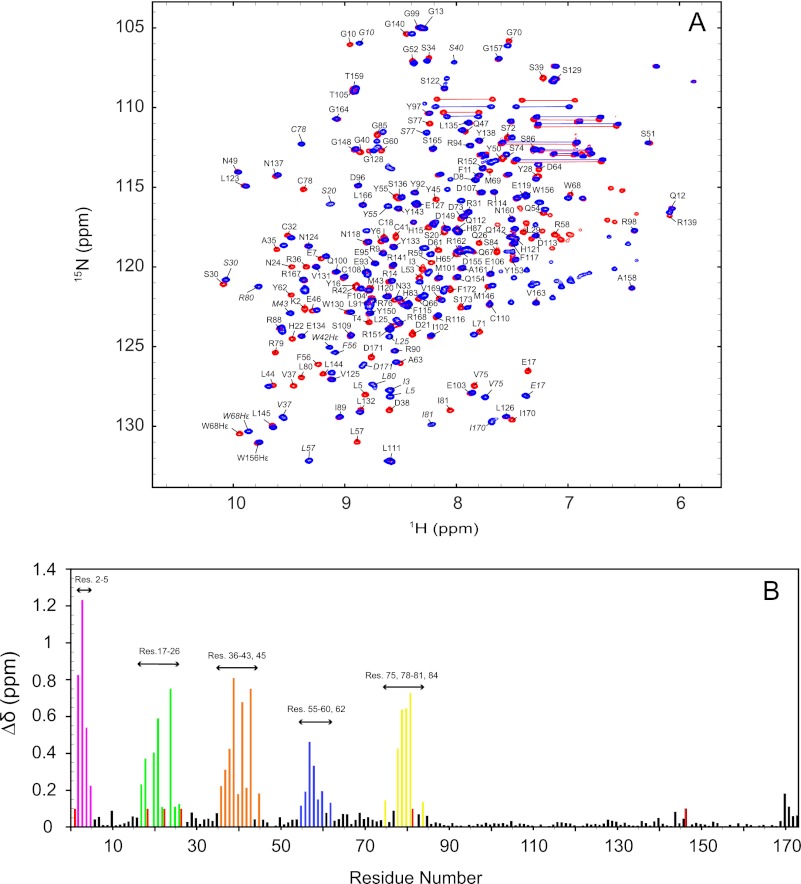
To assess the W42R mutant structure, we recorded 1H-15N HSQC spectra of the mutant and the wild-type protein under identical buffer conditions. Because backbone 1H-15N amide group resonances are very sensitive to conformation and electronic environment, 1H-15N HSQC spectra can be used as a fingerprint of the conformation of a protein. In the W42R mutant spectrum, 166 of 168 possible amide resonances were assigned. As evidenced by the superposition of the 1H-15N HSQC spectra of W42R and HGD shown in Fig. 1A, the mutant exhibits equally well dispersed resonances of uniform line widths as the wild-type protein, with most resonances overlapping or shifted only by a small amount. This suggests that the structure of the mutant is very similar to that of HGD. A quantitative comparison of the backbone chemical shifts between W42R and HGD reveals that all effects of the mutation are confined to the N-td, and perturbed resonances belong to amino acids close in either sequence or structure to the site of mutation (Fig. 1B). The largest combined amide 1H-15N chemical shift differences are associated with Ile-3, Ser-39, Cys-41, and Ile-81, with a maximum value of ∼1.25 ppm. As expected, removal of the tryptophan side chain is expected to affect amide resonances of residues located above or below the large aromatic ring because ring current shifts will no longer be present in the mutant. Indeed, such changes are observed for Arg-2–Lys-5, Val-75, Cys-78–Ile-81, and Ser-84.
FIGURE 1.
1H-15N HSQC spectra and chemical shift differences between the W42R mutant and HGD. A, superposition of the 1H-15N HSQC spectra of ∼0.1 mm W42R (blue contours) and ∼1 mm HGD (red contours) at 25 °C. 166 of 168 amide resonances were assigned and are labeled with amino acid name and number. HGD amide resonances are labeled in italic. B, combined amide 1H, 15N chemical shift differences between the W42R mutant and HGD versus residue number (Res.). Unassigned resonances are given an arbitrary value of 0.1 ppm.
At first sight, it seemed surprising that the replacement of the large hydrophobic side chain (Trp) by a long, positively charged side chain (Arg) did not cause a significant structural change. Given that in the HGD structure the Trp residue is positioned in the hydrophobic core of the N-td, one would have assumed that the positive charged Arg side chain would perturb the structure considerably. However, this appeared not to be the case.
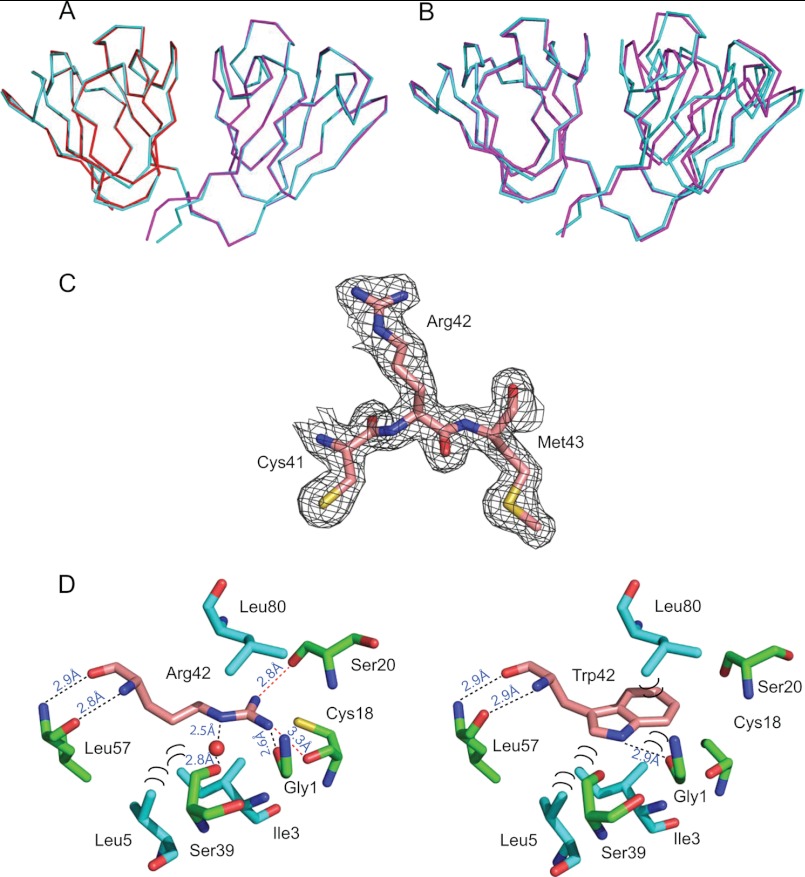
X-ray Structure
Conformation of our NMR findings was obtained by crystallography. Superposition of the N-td (residues 1–81) and C-td (residues 89–171) domains of the W42R mutant (Fig. 2A, colored red and magenta, respectively) onto those of HGD (Fig. 2A, colored cyan) yielded average pairwise r.m.s.d. values of 0.363 and 0.280 Å, respectively, for the backbone atoms. These low r.m.s.d. values confirm that the domain structures of the W42R mutant and HGD are extremely similar (Fig. 2A). Note that the two molecules of W42R in the asymmetric unit exhibit an average pairwise backbone atom r.m.s.d. value of 0.205 Å.
FIGURE 2.
Crystal structure of human W42R γD-crystallin. A, superposition of the N-td (red, residues 1–81) and C-td (magenta, residues 82–171) coordinates of W42R onto those of full-length HGD (cyan, PDB ID: 1HK0). B, best-fitting the W42R N-td-only coordinates (magenta, residues 1–81) onto full-length HGD (cyan, PDB ID: 1HK0). C, the electron density of residue Arg-42 in W42R, contoured at 1.0 electron density standard deviation. D, contacts around the Arg-42 side chain in the W42R mutant structure (left) and around Trp-42 in the HGD structure (PDB ID: 1HK0, right). Hydrogen bonds are labeled with black dots, and hydrophobic contacts are labeled with black semicircles.
Quite surprisingly, superposition of the overall W42R mutant and HGD structures (residues 1–171) yielded a somewhat larger, average backbone r.m.s.d. value of 0.795 Å. Indeed, the relative orientation between the N-td and C-td in the W42R mutant and HGD is slightly different with a tilt angle of 9.1° between the domains (Fig. 2B). This either could be caused by the Trp-Arg substitution or may simply be the result of crystal packing. Further detailed analysis of the W42R and the HGD structures revealed that there is some minor structural readjustment around the Arg side chain, involving hydrogen bonding and hydrophobic interactions, as detailed below.
The final refined electron density map of the W42R mutant reveals well defined electron density for the Arg residue (Fig. 2C). Surprisingly, a similar positioning of the Arg side chain within the hydrophobic core of the N-td to that of the Trp residue in the HGD structure is observed (Fig. 2D). Several interactions of the Arg residue with the surrounding atoms are identical to those seen for the Trp side chain, with several minor differences as detailed below. The backbone amide and carbonyl groups of Arg-42 form hydrogen bonds with the carbonyl and amide groups of Leu-57, respectively, and these are identical in wild-type HGD. Hydrogen bonds are formed between the Hϵ of the Arg-42 side chain and the backbone carbonyl oxygen atom of Ser-39, mediated by a buried water molecule inside the cavity, between one of the Arg-42 guanidino Hη atoms and the carbonyl group of Gly-1 and between the other Arg-42 guanidino Hη and the carbonyl group of Ser-20. There is only one hydrogen bond formed by the corresponding Trp-42 residue in the HGD structure (PDB ID: 1HK0), namely between the Hϵ of Trp-42 side chain and the backbone carbonyl group of Gly-1. Hydrophobic interactions involve the aliphatic portion of the Arg-42 side chain (Hβ, Hγ, and Hδ) and the side chains of Ile-3 and Leu-5 (Fig. 2D). In the HGD structure, the side chains of Ile-3 and Leu-5 also form similar hydrophobic contacts with the aromatic ring of the Trp-42 side chain. However, the hydrophobic contact between the side chain of Leu-80 and the six-membered ring of the Trp-42 side chain in the HGD structure is no longer observed in the W42R mutant because the guanidino group of the Arg-42 side chain occupies that position.
The presence of a buried water molecule inside the cavity of W42R is intriguing. As described above, it forms two hydrogen bonds: first, to the amide Hϵ of the Arg-42 side chain and, second, to the backbone carbonyl oxygen atom of Ser-39, bridging Arg-42 and Ser-39. In addition to these hydrogen bonds, it is also engaged in hydrogen bonding to the backbone carbonyl oxygen atom of Gly-40. In the HGD structure, such a water molecule is not present, and the backbone carbonyl oxygen of Gly-40 is directly hydrogen-bonded to the amide proton of Arg-59. In the W42R structure, however, the amide proton of Arg-59 is now hydrogen-bonded to the carbonyl oxygen atom (Oδ2) of the Asp-171 side chain. Thus, the additional contact from the Arg-59 backbone to Asp-171 located in the C-td of the protein appears to pull the C-terminal tail closer to the bottom of the N-td domain, causing the top part of the C-td to be slightly tilted away relative to the top of the N-td (Fig. 2B).
Whether the above interaction is responsible for the slightly different domain orientation or whether it is caused by a bound phosphate ion located between the two protein molecules in the crystal of the W42R mutant cannot be established unambiguously. This phosphate is coordinated by the side chains of Arg-59 and Arg-168 from each polypeptide, with each of the four oxygen atoms held by one guanidino group.
Because solubility of globular proteins may be associated with the extent of protein-protein interactions in solution, we carefully assessed whether the Arg-42 substitution could cause changes in the solvent-accessible surface of the protein. However, both the wild-type HGD and the W42R structures possess essentially identical solvent-accessible surface areas (8894.32 and 8546.23 Å2, respectively, as calculated by VMD (40), or 8669.40 and 8520.40 Å2, respectively, as calculated by NACCESS (41)). Therefore, no features of the globular structure of the W42R mutant protein can explain its involvement in cataract formation, and other properties needed to be considered and examined.
Equilibrium Unfolding of W42R Mutant
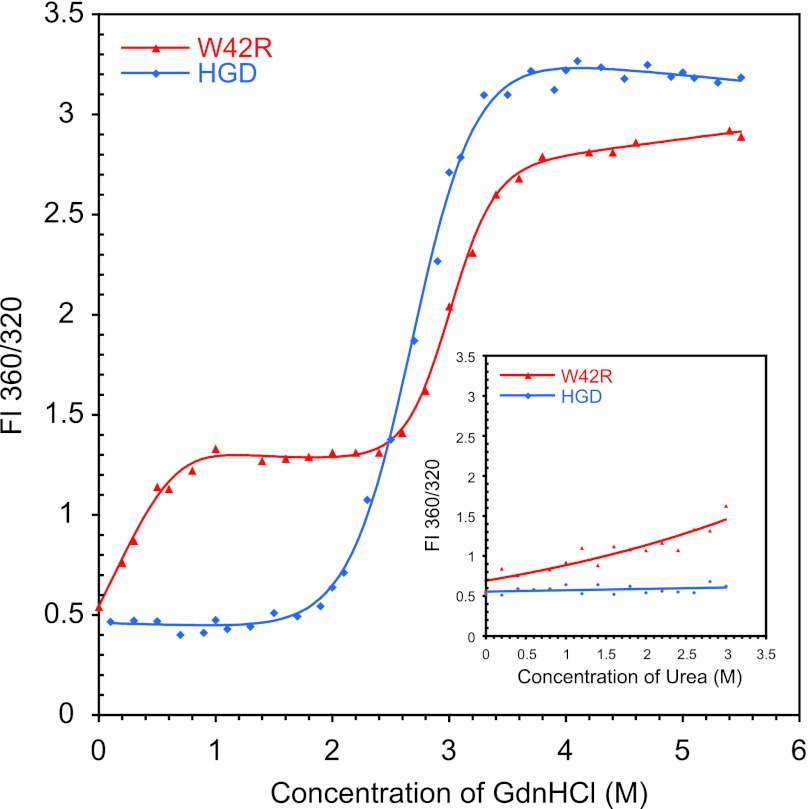
Because there were no noticeable changes in the structure that could explain the low solubility of the mutant, we analyzed the thermodynamic stability of W42R and compared this with HGD under close to physiological conditions (37 °C, pH 7.0). We performed equilibrium unfolding/refolding experiments and monitored the denaturation by tryptophan fluorescence. The unfolding curve of W42R clearly shows two-step unfolding (Fig. 3), indicating that a stable, highly populated unfolding intermediate exists. The first unfolding transition occurs with a GdnHCl half-point of 0.19 m followed by a prominent plateau throughout the interval of GdnHCl concentrations from 1.2–2.4 m (Fig. 3). The second transition exhibits a GdnHCl half-point of 3.1 m. The presence of an intermediate was previously observed for the Q54E mutant of HGD, for which it was shown that the unfolded portion of the protein comprised the N-td, whereas the C-td remained in a native-like structure (22). The unfolding isotherm for W42R was fit to a three-state model. This yielded a ΔG0N-I of 2.4 ± 1.1 kcal/mol for the first transition and ΔG0I-U = 9.8 ± 0.8 kcal/mol for the second transition. Under identical conditions, HGD unfolds with a midpoint at 3.1 m GdnHCl, and no intermediate state is observed. Therefore, the unfolding isotherm was fit to a two-state model with an apparent ΔG0N-U = 6.0 ± 0.3 kcal/mol. This demonstrates that the substitution of Trp-42 by Arg-42 results in a dramatic destabilization of the N-td, consistent with the location of the amino acid substitution.
FIGURE 3.
Denaturant induced unfolding of W42R (red) and HGD (blue) by GdnHCl or urea (inset) at 37 °C. All samples contained 10 μg/ml protein in 100 mm sodium phosphate buffer, pH 7.0, 5 mm DTT, 1 mm EDTA and GdnHCl from 0 to 5.5 m and urea from 0 to 3 m. FI, fluorescence intensity ratio.
Because the GdnHCl unfolding curve revealed that the W42R mutant was already starting to unfold at the lowest GdnHCl concentration employed, we repeated the unfolding experiment with urea as the chaotropic agent under identical conditions. These data are provided in the inset of Fig. 3. As can be seen, at low urea concentrations (<1 m), the W42R protein starts to unfold, whereas HGD displays no sign of unfolding up to 3 m urea. The above results demonstrate that although no significant changes in the overall three-dimensional structure of the W42R protein when compared with HGD are observed, dramatic stability changes can occur. In the context of γD-crystallins, this may well relate to the aggregation and precipitation properties of destabilizing mutants that are associated with cataract formation. Indeed, protein aggregation and precipitation prevented us from extracting accurate data for thermal denaturation; precipitation occurred prior to the completion of the unfolding, and therefore, unfolding was irreversible.
Protease Susceptibility of W42R
During purification of the W42R mutant protein, we noticed that degradation products were present, whereas this was not the case for HGD or other mutants. In particular, an ∼11-kDa species was observed, and its size was accurately determined by MALDI as 11,313.54 Da. Adding protease inhibitor mixture to the cell lysate, as well as including PMSF and EDTA in all purification buffers, suppressed the degradation. This suggested that the W42R mutant protein was particularly susceptible to proteases.
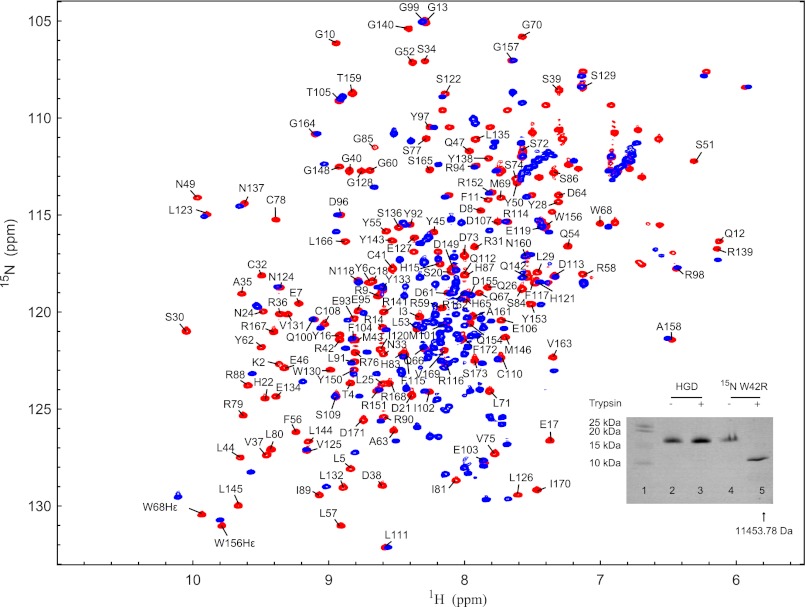
Therefore, we subjected intact W42R protein to trypsin cleavage and compared the mutant behavior with that of HGD. Overnight incubation of the W42R protein with trypsin generated an ∼11-kDa species (Fig. 4, inset, lanes 4 and 5), whereas no such band was observed with HGD under identical conditions. The molecular mass of the ∼11-kDa band from the 15N-labeled sample was determined by MALDI as 11,453.78 Da (when compared with 11,313.54 Da from the unlabeled sample; see above). These masses suggest that the final cleavage occurred between Arg-79 and Leu-80 because the calculated molecular masses for a fragment from Leu-80 to Ser-173 are 11,323.69 and 11,472.69 Da for natural abundance and 15N-labeled W42R, respectively. Comparison of the 1H-15N HSQC spectra prior to and after trypsin treatment shows that all resonances of N-td have disappeared, whereas those belonging to the C-td are present with the same chemical shifts (Fig. 4). Several smaller fragments (<2000 Da) were also detected by MALDI (data not show), and the 1H-15N HSQC spectrum of the trypsin-treated sample displays resonances in the random coil region, indicating that N-td is degraded into small peptides. These results indicate that W42R contains a small population of a species with an unfolded N-td that can be effectively proteolyzed by trypsin.
FIGURE 4.
Superposition of the 1H-15N HSQC spectra of untreated (red) and trypsin-digested (blue) W42R. Trypsin cleavage reactions were performed overnight at 37 °C in 20 mm sodium phosphate, pH 7.0, 5 mm DTT, 0.02% NaN3 with an enzyme-to-substrate ratio of 1:100 (w/w). A comparison between trypsin-digested wild-type HGD and the W42R mutant by SDS-PAGE is shown in the inset. Note that lane 1 contains protein standards, lanes 2 and 3 contain HGD in the absence and presence of trypsin, respectively, and lanes 4 and 5 contain W42R in the absence and presence of trypsin, respectively.
Unfolded Species Determined by NMR Spectroscopy
It has been hypothesized that cataracts may be caused by an off-pathway aggregation under (un)folding equilibrium conditions, and a few studies showed that γD-crystallin is prone to aggregation under partially denaturing conditions (42). Evidence for a partially folded species was obtained by NMR spectroscopy. 1H-15N HSQC spectra of W42R were recorded at 900 MHz with a large number of scans at 25, 37, and 42 °C. Several small, well resolved resonances that were not observed at 600 MHz were detected. For assignment of these resonances, this spectrum was compared with that of the V75D mutant of HGD (V75D) that is known to possesses an unfolded N-td and folded C-td at 3.6 m urea and 37 °C, as shown by fluorescence spectroscopy (15) and solution NMR spectroscopy.3 Comparing the 1H-15N HSQC spectra of native W42R, trypsin-digested W42R, HGD C-td, and V75D in 3.6 m urea, many minor resonances were assigned to residues that belong to the C-td (Fig. 5A).
FIGURE 5.
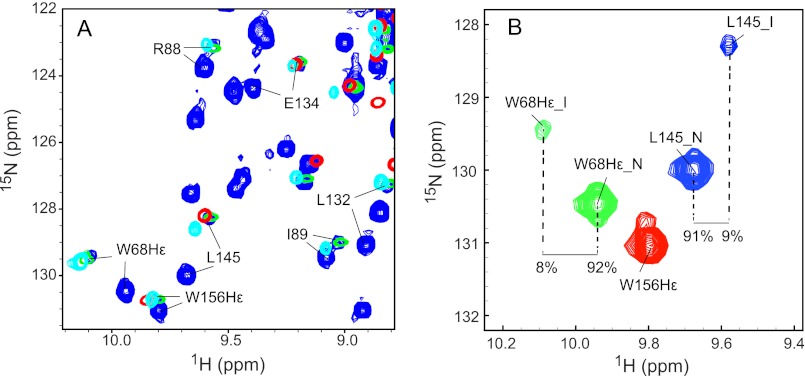
NMR evidence for the presence of a partially (un)folded species of W42R. A, expanded regions of the superposition of the 1H-15N HSQC spectra of W42R, trypsin-digested 15N W42R, HGD C-td, and V75D. Spectra of W42R were recorded at 900 MHz at 37 °C (blue contours), spectra of trypsin-digested 15N W42R were recorded at 800 MHz at 37 °C (green contours), spectra of HGD C-td under native conditions were recorded at 600 MHz at 25 °C (red contours), and spectra of V75D in 3.6 m urea were recorded at 900 MHz at 37 °C (cyan contours), respectively. B, expanded region of the 1H-15N HSQC spectrum of the W42R mutant protein at 900 MHz and 37 °C. The Trp-68 Nϵ1 resonances (green contours), Trp-156 Nϵ1 resonances (red contours), and Leu-145 amide resonances (blue contours) are displayed. Resonances associated with the natively folded protein are labeled with N, and resonances associated with the partially unfolded state are labeled with I.
To estimate the amount of the partially folded species of the W42R mutant under native conditions, we selected the Trp-68 Hϵ1 and Leu-145 amide resonances as representative resonances for the N-td and the C-td. The relative peak volumes in the 900-MHz 1H-15N HSQC spectrum at 37 °C for the natively folded, major state (N) of W42R and for the partially folded, minor state (I) (Fig. 5B) yielded 92:8 for the Trp-68 Hϵ1 resonance and 91:9 for the Leu-145 amide resonance. Given the identical percentages, it seems likely that these small resonances belong to the same chemical species. With increasing temperature, the amount of the I-state increased, implying that an unfolding process is involved.
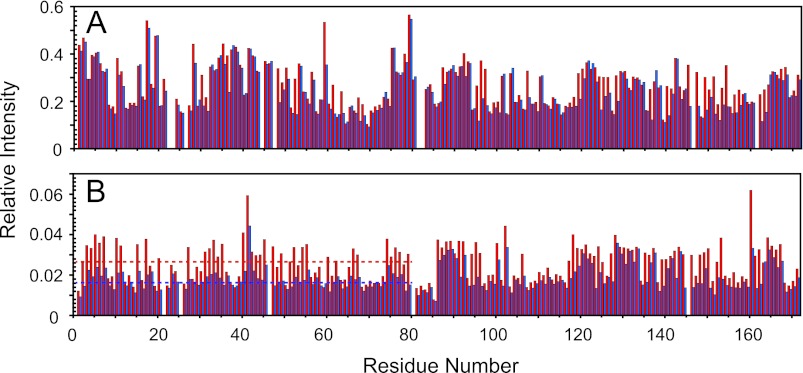
To further confirm that the small resonances do not arise from an unrelated chemical species, but belong to the partially folded minor I-state that can exchange with the major N-state, we also performed H/D exchange experiments for 15N-labeled HGD and the W42R variant. After the addition of 50% (v/v) D2O to the 15N-labeled samples, 1H-15N HSQC spectra were recorded as a function of time. In the folded state, slow exchange of amide hydrogens with deuterons occurs (43), whereas in the unfolded state, exchange is fast. Therefore, if any changes in H/D exchange for the folded state are observed over time, this would support the notion that chemical exchange between the partially folded and native state takes place. As shown in Fig. 6, the W42R protein exhibited pronounced H/D exchange of amide protons, throughout the entire N-td, whereas residues in the C-td displayed no detectable changes. Thus, the H/D exchange also supports the view that the I-state possesses an unfolded N-td and a folded C-td.
FIGURE 6.
H/D exchange of HGD and W42R mutant proteins. A, relative intensities of HGD resonances in 50% D2O to HGD resonances in 5% D2O at 0 h (red) and 30 h (blue). B, relative intensities of W42R resonances in 50% D2O to HGD resonances in 5% D2O at 0 h (red) and 30 h (blue). The average relative intensity of resonances in the 1H-15N HSQC spectra the N-td of W42R is indicated by the dotted line.
UV-C Irradiation
Given that Trp-42 appears to be important for protein stability and that the loss of this residue leads to aggregate formation, possibly causing cataract, we probed whether the loss of a Trp could be associated with photodamage of HGD. UV is an environmental factor that damages tryptophans, and Trp-42 is the most UV-sensitive tryptophan in γD-crystallin (44). Covalent damage of Trp residues leads to ring cleavage, originating from a short-lived Trp cation radical (45, 46) that, upon photooxidation, turns to kynurenine or derivatives. Therefore, the mutation of tryptophan to arginine in the W42R mutant may serve a model for photodamaged γD-crystallin and UV-induced cataract. To evaluate whether UV damage in the tryptophans of HGD results in similar properties as seen in the W42R mutant, we exposed HGD to different doses of UV.
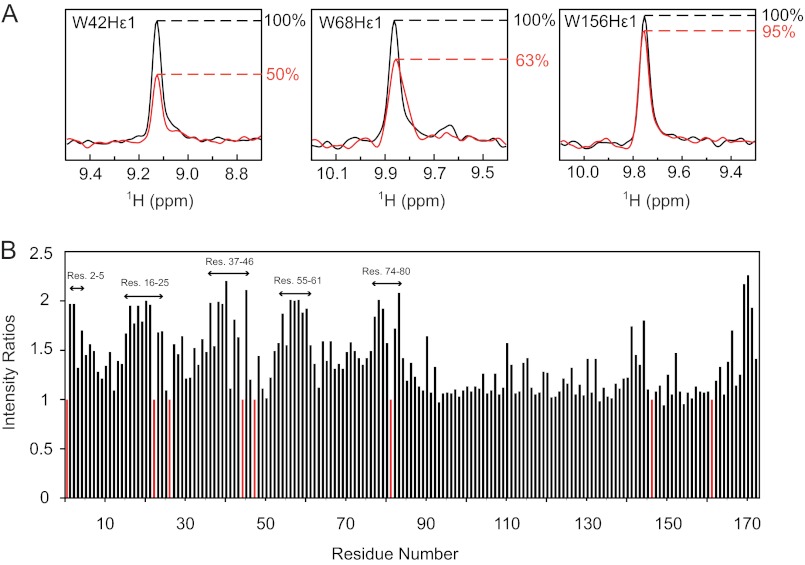
15N-labeled wild-type protein was exposed to UV-C (254 nm) light, and 1H-15N HSQC spectra were recorded before and after irradiation with increasing energy doses until new resonances appeared (data not show). After irradiation, the intensity of the Trp-42 Hϵ1 resonance was reduced by 50%, that of Trp-68 was reduced by 37%, whereas that of Trp-156 was only decreased by 5% (Fig. 7A). Due to overlap of Trp-130 Hϵ1 and Phe-115 amide resonances, the intensity decrease of the Trp-130 Hϵ1 resonance could not be calculated reliably. Overall, many new resonances appeared in the 1H-15N HSQC spectrum after irradiation, indicating that new protein species had appeared. We estimated which parts (amino acids) of HGD experienced damage by measuring the intensities of resonances that were associated with undamaged protein over the time course of the irradiation. These data for a UV exposure <1 J/cm2 are provided in Fig. 7B. The largest changes occurred in resonances belonging to residues in N-td, with changes observed for resonances of amino acids 2–5, 16–25, 37–46, 55–61, and 74–80. Some changes were also seen in the C-td, namely for residues ∼144 and 170–173. Interestingly, the UV-affected regions are essentially identical to those for which differences in the spectra between the W42R mutant and HGD were noted (Fig. 1). This suggests that the changes observed in the irradiated HGD spectrum were caused by damage of Trp-42, rather than by damage to all the residues that experience changes.
FIGURE 7.
Effect of UV exposure on 15N HGD. A, slices through the tryptophan Hϵ1 resonances in the 1H-15N HSQC spectra of UV-exposed (red) versus nonexposed (black) protein are superimposed. B, intensity ratios of native resonances of 15N HGD to those of 15N HGD after UV exposure.
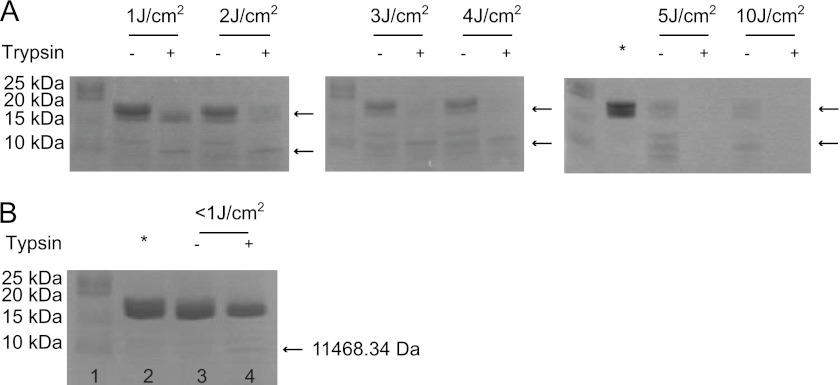
We further investigated whether the Trp-42-damaged HGD protein behaved similar to the W42R mutant protein in the trypsin digestion experiment. Not surprisingly, the HGD protein became susceptible to degradation by trypsin after having been irradiated at 254 nm (Fig. 8A). The trypsin digestion product (indicated by the lower arrow on the SDS-PAGE gel) was also analyzed by MALDI, and this fragment had a molecular mass of 11,464.82 Da, very similar (∼120-Da difference) to that found for the trypsin-generated W42R C-td fragment. The UV-irradiated 15N HGD NMR sample was also subjected to trypsin cleavage and analyzed by SDS-PAGE as well as MALDI, revealing that a fragment of 11,468.34 Da is generated (Fig. 8B) whose molecular mass is in agreement with the previously observed and predicted mass of a 15N-labeled fragment, comprising residues Leu-80 to Ser-173. Although the native HGD protein is resistant to trypsin digestion (Fig. 4), UV irradiation appears to damage Trp-42, as shown by NMR, and initiates the formation of a partially folded species that is easily cleaved by protease. At UV-C doses above 5 J/cm2, the irradiated HGD was completely digested by trypsin, suggesting more widespread damage also to the C-td.
FIGURE 8.
UV-C exposure and trypsin digestion of HGD and 15N HGD. A and B, SDS-PAGE of UV-C-exposed and trypsin-digested natural abundance HGD for increasing energy doses (1.0, 2.0, 3.0, 4.0, 5.0 J/cm2) (A) and UV-C-exposed and trypsin-digested 15N HGD (B). In B: lane 1, protein standard; lane 2, 15N HGD; lane 3, 15N HGD incubated at 37 °C overnight without trypsin; lane 4, 15N HGD incubated at 37 °C overnight with trypsin. Nonexposed HGD samples are loaded in the lanes marked by asterisks.
UV irradiation appears to cause other changes in the protein as well because increased molecular masses were observed for the HGD protein, namely 20,559.77 Da (1 J/cm2), 20,736.57 Da (2 J/cm2), 21,197.66 Da (3 J/cm2), 21,630.29 Da (4 J/cm2), and 21,525.03 Da (5 J/cm2). All the above findings strongly suggest that UV-mediated protein damage, in particular Trp-42 oxidation, plays a critical role in destabilizing γD-crystallin, contributing to the etiology of age-related cataract.
DISCUSSION
Many proteins are associated with deposition diseases in humans, including Alzheimer disease, prion-related Creutzfeldt-Jakob disease, type II diabetes, and cataract. The proteins involved exhibit a common feature: the presence of a partially unfolded or non-native conformation (47–50). Elucidating how these proteins lose their structure by environmental factors is crucial for understanding the pathology of protein deposition diseases. One of the prevailing models for cataract formation evokes off-pathway folding and aggregation. In this model, unfolded protein escapes from sequestration by α-crystallin and forms aggregates. Support for this model is based on results with hydrophobic core mutants of γD-crystallin under denaturing conditions (15). Although suggestive, there was no direct evidence linking protein damage to aggregation under physiological conditions up to now.
Our current research appears to supply this missing link since it revealed that the mutant protein contains a population of partially folded protein, although no significant difference in the overall structure from HGD can be discerned for the major conformer. At 37 °C, a noticeable amount (∼10%) of partially folded species is present, similar to related observations for a F9S mutant of mouse γS-crystallin that is associated with Opj cataract in mice and for which an unfolding intermediate was found to be populated to ∼1–2% at physiological conditions (19, 20).
It is also worth noting that the structures of other HGD mutants, such as R36S and R58H (51, 52), do not exhibit major structural changes when compared with wild type. As to causes for their solubility changes, for the R36S mutant, it has been suggested that the elimination of the surface charge by the substitution of the Arg-36 side chain by Ser induces the mutant protein to engage in protein-protein contacts that are not possible in the wild-type protein (52). Similarly, for the R58H mutant, it has been proposed that the reduced solubility is mainly due to an effect of the mutation on the solution phase (51). Altogether, our current results and combined previous data clearly show that no major conformational changes of HGD are necessary, as evidenced by the very similar x-ray structures, but that it is solution behavior and in particular (un)folding dynamics of the different mutants that play critical roles in cataract formation. Indeed, this notion is unambiguously borne out by the results of the study presented here.
Most importantly, our findings link the behavior of the cataract-associated W42R mutant to UV-damaged HGD. Although effects of UV damage on crystallins have been studied previously (44), no biophysical data about the structural consequences have been analyzed. Here we investigated whether the W42R mutant protein and UV-damaged HGD share common properties. Our NMR data revealed that the UV-induced damage was confined to the area around Trp-42, essentially limited to the N-td, supporting previous observations on the susceptibility of this Trp-42 residue upon UV exposure (53).
The combined data on the W42R mutant and UV-irradiated HGD demonstrate that modification of even a single residue is sufficient to destabilize γD-crystallin enough to induce protein aggregation in vitro, even if the x-ray structure of the mutant is essentially identical to that of HGD.
Acknowledgments
F. J. thanks advisor Prof. Yongming Bao at the Dalian University of Technology for training and support. We thank Mike Delk for NMR technical support and Alexander M. J. J. Bonvin for calculating solvent-accessible surface area using the program NACCESS.
This work was supported, in whole or in part, by National Institutes of Health Grant EY021193 (to A. M. G). This work was also supported by a scholarship from the China Scholarship Council (to F. J.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 4GR7) have been deposited in the Protein Data Bank (http://wwpdb.org/).
J. Jung, unpublished data.
- HGD
- human γD-crystallin
- N-td
- N-terminal domain
- C-td
- C-terminal domain
- GdnHCl
- guanidinium hydrochloride
- HSQC
- heteronuclear single quantum correlation
- r.m.s.d.
- root mean square deviation
- H/D
- hydrogen/deuterium.
REFERENCES
- 1. Reddy M. A., Bateman O. A., Chakarova C., Ferris J., Berry V., Lomas E., Sarra R., Smith M. A., Moore A. T., Bhattacharya S. S., Slingsby C. (2004) Characterization of the G91del CRYBA1/3-crystallin protein: a cause of human inherited cataract. Hum. Mol. Genet. 13, 945–953 [DOI] [PubMed] [Google Scholar]
- 2. Vijaya R., Gupta R., Panda G., Ravishankar K., Kumaramanickavel G. (1997) Genetic analysis of adult-onset cataract in a city-based ophthalmic hospital. Clin. Genet. 52, 427–431 [DOI] [PubMed] [Google Scholar]
- 3. Rahi J. S., Dezateux C. (2000) Congenital and infantile cataract in the United Kingdom: underlying or associated factors. Invest. Ophthalmol. Vis. Sci. 41, 2108–2114 [PubMed] [Google Scholar]
- 4. Lampi K. J., Ma Z., Shih M., Shearer T. R., Smith J. B., Smith D. L., David L. L. (1997) Sequence analysis of βA3, βB3, and βA4 crystallins completes the identification of the major proteins in young human lens. J. Biol. Chem. 272, 2268–2275 [DOI] [PubMed] [Google Scholar]
- 5. Hejtmancik J. F. (2008) Congenital cataracts and their molecular genetics. Semin. Cell Dev. Biol. 19, 134–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andley U. P. (2007) Crystallins in the eye: Function and pathology. Prog. Retin. Eye Res. 26, 78–98 [DOI] [PubMed] [Google Scholar]
- 7. Shiels A., Bennett T. M., Hejtmancik J. F. (2010) Cat-Map: putting cataract on the map. Mol. Vis. 16, 2007–2015 [PMC free article] [PubMed] [Google Scholar]
- 8. Oyster C. W. (1999) The Human Eye: Structure and Function, Sinauer Associates, Sunderland, MA [Google Scholar]
- 9. Sinha D., Wyatt M. K., Sarra R., Jaworski C., Slingsby C., Thaung C., Pannell L., Robison W. G., Favor J., Lyon M., Wistow G. (2001) A temperature-sensitive mutation of Crygs in the murine Opj cataract. J. Biol. Chem. 276, 9308–9315 [DOI] [PubMed] [Google Scholar]
- 10. Sinha D., Esumi N., Jaworski C., Kozak C. A., Pierce E., Wistow G. (1998) Cloning and mapping the mouse Crygs gene and non-lens expression of γS-crystallin. Mol Vis 4, 8. [PubMed] [Google Scholar]
- 11. Everett C. A., Glenister P. H., Taylor D. M., Lyon M. F., Kratochvilova-Loester J., Favor J. (1994) Mapping of six dominant cataract genes in the mouse. Genomics 20, 429–434 [DOI] [PubMed] [Google Scholar]
- 12. Graw J., Löster J., Soewarto D., Fuchs H., Reis A., Wolf E., Balling R., Hrabé de Angelis M. (2002) V76D mutation in a conserved γD-crystallin region leads to dominant cataracts in mice. Mamm. Genome 13, 452–455 [DOI] [PubMed] [Google Scholar]
- 13. Graw J., Neuhäuser-Klaus A., Klopp N., Selby P. B., Löster J., Favor J. (2004) Genetic and allelic heterogeneity of Cryg mutations in eight distinct forms of dominant cataract in the mouse. Invest. Ophthalmol. Vis. Sci. 45, 1202–1213 [DOI] [PubMed] [Google Scholar]
- 14. Flaugh S. L., Kosinski-Collins M. S., King J. (2005) Contributions of hydrophobic domain interface interactions to the folding and stability of human γD-crystallin. Protein Sci. 14, 569–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moreau K. L., King J. (2009) Hydrophobic core mutations associated with cataract development in mice destabilize human γD-crystallin. J. Biol. Chem. 284, 33285–33295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flaugh S. L., Kosinski-Collins M. S., King J. (2005) Interdomain side-chain interactions in human γD crystallin influencing folding and stability. Protein Sci. 14, 2030–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kosinski-Collins M. S., Flaugh S. L., King J. (2004) Probing folding and fluorescence quenching in human γD crystallin Greek key domains using triple tryptophan mutant proteins. Protein Sci. 13, 2223–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fu L., Liang J. J. (2002) Conformational change and destabilization of cataract γC-crystallin T5P mutant. FEBS Lett. 513, 213–216 [DOI] [PubMed] [Google Scholar]
- 19. Lee S., Mahler B., Toward J., Jones B., Wyatt K., Dong L., Wistow G., Wu Z. (2010) A single destabilizing mutation (F9S) promotes concerted unfolding of an entire globular domain in γS-crystallin. J. Mol. Biol. 399, 320–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mahler B., Doddapaneni K., Kleckner I., Yuan C., Wistow G., Wu Z. (2011) Characterization of a transient unfolding intermediate in a core mutant of γS-crystallin. J. Mol. Biol. 405, 840–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen J., Callis P. R., King J. (2009) Mechanism of the very efficient quenching of tryptophan fluorescence in human γD- and γS-crystallins: the γ-crystallin fold may have evolved to protect tryptophan residues from ultraviolet photodamage. Biochemistry 48, 3708–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen J., Flaugh S. L., Callis P. R., King J. (2006) Mechanism of the highly efficient quenching of tryptophan fluorescence in human γD-crystallin. Biochemistry 45, 11552–11563 [DOI] [PubMed] [Google Scholar]
- 23. Chen J., Toptygin D., Brand L., King J. (2008) Mechanism of the efficient tryptophan fluorescence quenching in human γD-crystallin studied by time-resolved fluorescence. Biochemistry 47, 10705–10721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Searle B. C., Dasari S., Wilmarth P. A., Turner M., Reddy A. P., David L. L., Nagalla S. R. (2005) Identification of protein modifications using MS/MS de novo sequencing and the OpenSea alignment algorithm. J. Proteome Res. 4, 546–554 [DOI] [PubMed] [Google Scholar]
- 25. Robman L., Taylor H. (2005) External factors in the development of cataract. Eye 19, 1074–1082 [DOI] [PubMed] [Google Scholar]
- 26. Wang B., Yu C., Xi Y. B., Cai H. C., Wang J., Zhou S., Zhou S., Wu Y., Yan Y. B., Ma X., Xie L. (2011) A novel CRYGD mutation (p.Trp43Arg) causing autosomal dominant congenital cataract in a Chinese family. Hum. Mutat. 32, E1939–E1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jung J., Byeon I. J., Wang Y., King J., Gronenborn A. M. (2009) The structure of the cataract-causing P23T mutant of human γD-crystallin exhibits distinctive local conformational and dynamic changes. Biochemistry 48, 2597–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ji F., Jung J., Gronenborn A. M. (2012) Structural and biochemical characterization of the childhood cataract-associated R76S mutant of human γD-crystallin. Biochemistry 51, 2588–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kantardjieff K. A., Rupp B. (2003) Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 12, 1865–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pflugrath J. W. (1999) The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 31. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 32. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 35. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 37. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 38. Keller B., Christen M., Oostenbrink C., van Gunsteren W. F. (2007) On using oscillating time-dependent restraints in MD simulation. J. Biomol. NMR 37, 1–14 [DOI] [PubMed] [Google Scholar]
- 39. Goddard T. D., Kneller D. G. (2004) SPARKY 3, 3.110 Ed., University of California, San Francisco [Google Scholar]
- 40. Humphrey W., Dalke A., Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 41. Hubbard S. J., Thornton J. M. (1993) NACCESS, Department of Biochemistry and Molecular Biology, University College London [Google Scholar]
- 42. Kosinski-Collins M. S., King J. (2003) In vitro unfolding, refolding, and polymerization of human γD crystallin, a protein involved in cataract formation. Protein Sci. 12, 480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wüthrich K., Wagner G. (1979) Nuclear magnetic resonance of labile protons in the basic pancreatic trypsin inhibitor. J. Mol. Biol. 130, 1–18 [DOI] [PubMed] [Google Scholar]
- 44. Wang S. S., Wen W. S. (2010) Examining the influence of ultraviolet C irradiation on recombinant human γD-crystallin. Mol. Vis. 16, 2777–2790 [PMC free article] [PubMed] [Google Scholar]
- 45. Bryant F. D., Santus R., Grossweiner L. I. (1975) Laser flash photolysis of aqueous tryptophan. J. Phys. Chem. 79, 2711–2716 [Google Scholar]
- 46. Bent D. V., Hayon E. (1975) Excited state chemistry of aromatic amino acids and related peptides. III. Tryptophan. J. Am. Chem. Soc. 97, 2612–2619 [DOI] [PubMed] [Google Scholar]
- 47. Chiti F., Dobson C. M. (2006) Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 [DOI] [PubMed] [Google Scholar]
- 48. Dobson C. M. (2004) Principles of protein folding, misfolding and aggregation. Semin. Cell Dev. Biol. 15, 3–16 [DOI] [PubMed] [Google Scholar]
- 49. Nicholson E. M., Mo H., Prusiner S. B., Cohen F. E., Marqusee S. (2002) Differences between the prion protein and its homolog Doppel: a partially structured state with implications for scrapie formation. J. Mol. Biol. 316, 807–815 [DOI] [PubMed] [Google Scholar]
- 50. Mitraki A., King J. (1989) Protein folding intermediates and inclusion body formation. Nature Biotechnology 7, 690–697 [Google Scholar]
- 51. Basak A., Bateman O., Slingsby C., Pande A., Asherie N., Ogun O., Benedek G. B., Pande J. (2003) High-resolution X-ray crystal structures of human γD crystallin (1.25 Ä) and the R58H mutant (1.15 Ä) associated with aculeiform cataract. J. Mol. Biol. 328, 1137–1147 [DOI] [PubMed] [Google Scholar]
- 52. Kmoch S., Brynda J., Asfaw B., Bezouska K., Novák P., Rezácová P., Ondrová L., Filipec M., Sedlácek J., Elleder M. (2000) Link between a novel human γD-crystallin allele and a unique cataract phenotype explained by protein crystallography. Hum. Mol. Genet. 9, 1779–1786 [DOI] [PubMed] [Google Scholar]
- 53. Tallmadge D. H., Borkman R. F. (1990) The rates of photolysis of the four individual tryptophan residues in UV exposed calf γ-II crystallin. Photochem. Photobiol. 51, 363–368 [DOI] [PubMed] [Google Scholar]