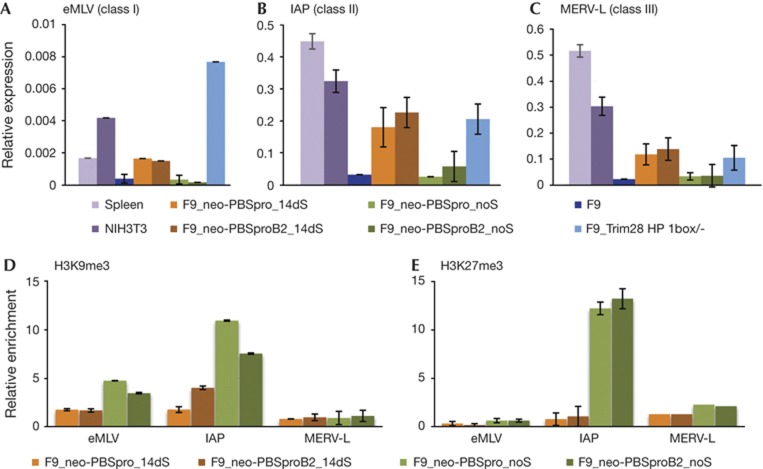
Figure 5.
Expression pattern and chromatin modifications of classes I, II and III ERVs. Quantitative RT–PCR analysis of (A) eMLVs (class I), (B) IAP (class II) and (C) MERV-L (class III) messenger RNA expression in embryonic and differentiated cells. Bars represent the mean±s.e.m. of three independently prepared samples, relative to three control genes (UBC, CYCA and Gapdh). ChIP-based measurement of (D) H3K9me3 and (E) H3K27me3 at the viral sequence of cells infected with MLV containing neor with PBSpro or PBSproB2 and selected for 14 days or infected without selection. Relative enrichment values (% of input) in all ChIP experiments were normalized to the relative enrichment at the Gapdh promoter. Negative and positive control genes gave the expected enrichment (not shown). ChIP, chromatin immunoprecipitation; MLV, murine leukaemia virus; ERV, endogenous retrovirus; PBSpro, proline tRNA primer-binding site; RT–PCR, reverse transcription PCR.