Figure 5.
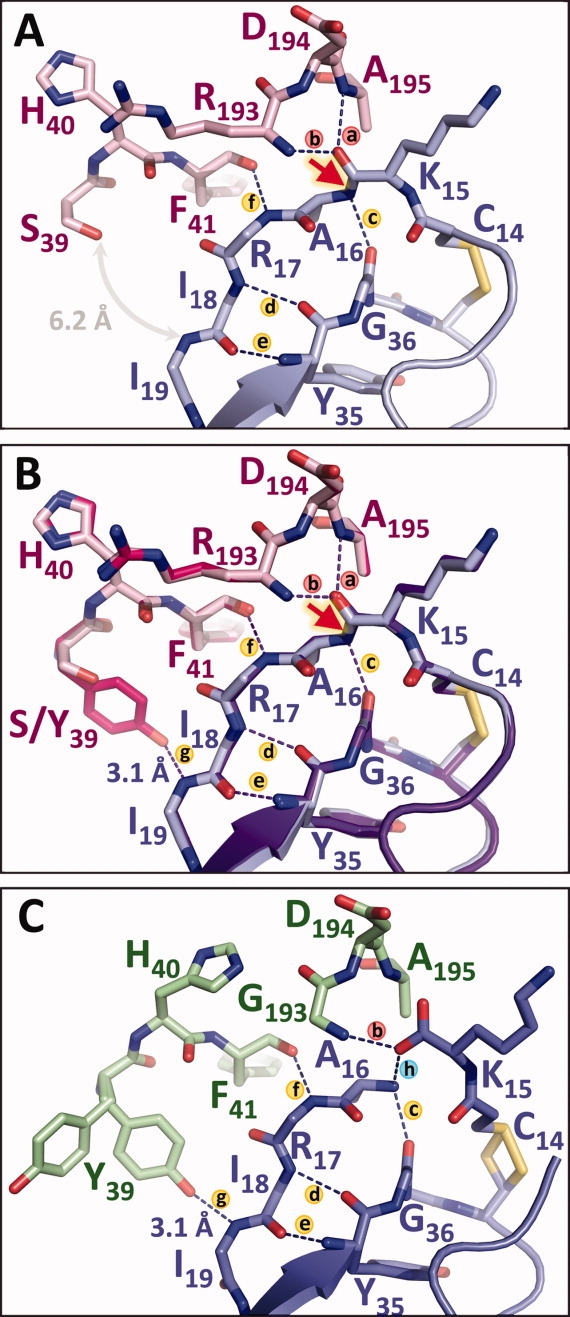
Primed-side H-bonds stabilize trypsin-inhibitor interactions and slow inhibitor hydrolysis. (A) In the mesotrypsin·BPTI structure (PDBID 2RP9), H-bonds (a and b) represent the “oxyanion hole” that will stabilize negative charge on the carbonyl oxygen during the cleavage of the scissile bond (red arrow). H-bonds (c–e) to the inhibitor scaffold and (f) to the enzyme counteract leaving group dissociation, but the absence of Tyr-39 leaves a large gap that may facilitate leaving group dissociation. (B) Mutation of mesotrypsin Ser-39 to Tyr restores H-bond g that is present in other trypsins, stabilizing the P3′ and P4′ residues between the enzyme and the inhibitor scaffold. (C) The structure of rat anionic trypsin bound to cleaved BPTI* (green/periwinkle; PDBID 3FP7) illustrates how H-bonds (c–g) may function cooperatively in retention of the primed-side leaving group after scissile bond cleavage. The opening of Tyr-39 seen in the alternate conformational rotamer in this structure may reflect the movement required of this residue prior to leaving group dissociation.
