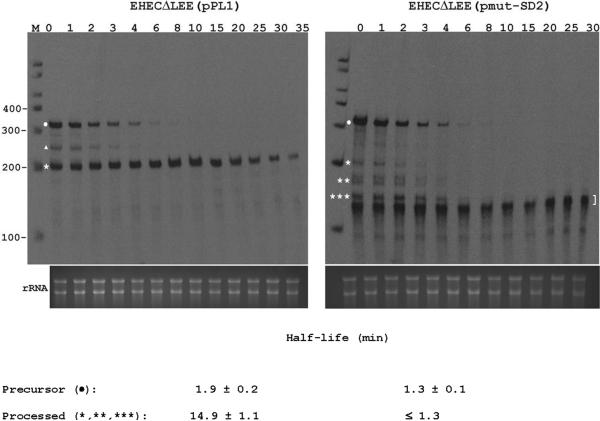
Figure 6. Effect of SD2 disruption on the decay of espAD mRNA.
EHECΔLEE was transformed with pPL1 (wild-type espAD) or its derivative pmut-SD2 (espAD with a mutated SD2 element (see Fig 3A)). Exponential-phase cultures were treated with rifampicin to inhibit transcription. RNA was isolated at time intervals (minutes after addition of rifampicin are shown at the top of each panel) and analyzed by RPA. Precursor mRNA is marked with a dot, while processed mRNA species are marked by a triangle and asterisks. A bracket denotes partially digested probe. An aliquot of each hybridization reaction was also examined by gel electrophoresis and ethidium staining (bottom of each panel). Mean half-life values ± SD are shown. The upper limit on the half-life of the mutant processed species (≤ 1.3 min for *, **, and *** derived from pmut-SD2) is based on the observation that these intermediates disappeared from rifampicin-treated cells at the same rate as their precursor, which decayed with a half-life of 1.3 ± 0.1 min.