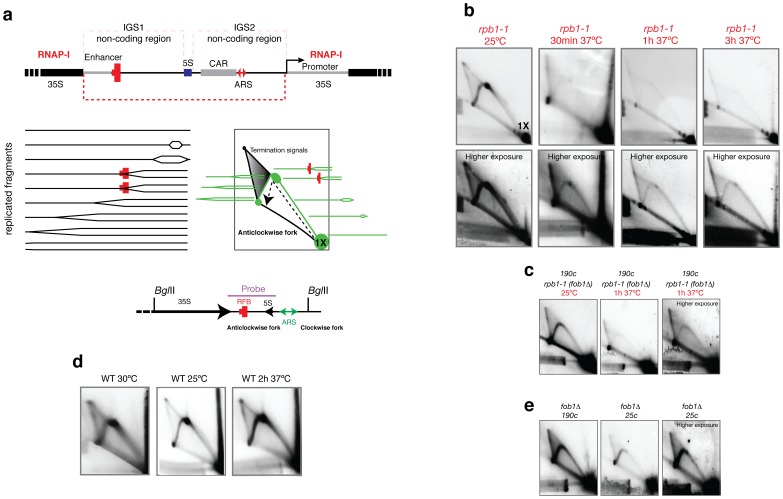
Figure 1. The replication of rDNA is affected in the absence of the largest subunit of RNAP-II.
(a) Diagram of the rDNA with the locations of the replication barrier (RFB), replication origin (ARS) and cohesin binding sequences (CAR). Below, the theoretical schemes for the two-dimensional agarose gel electrophoresis of chromatin digested with BglII are depicted. The accumulation of RIs at the RFB is expected at 1.49X. 1X represents the position of the linear, unreplicated fragment. The hybridisation probe used for the Southern protocol and hybridisation was amplified by PCR and column purified (Qiagen). The amplified fragment is represented above. The direction of the anticlockwise and clockwise replication fork is shown below. (b) Results obtained after the isolation of DNA from asynchronous cultures growing exponentially at different temperatures. Higher exposure images are shown below. Significantly fewer amounts of RIs were detected in the absence of RPB1 (rpb1–1 ts strain). 1X signals were similar among the 4 experiments (25°C, 37°C for 30 minutes, 37°C for 1 hour and 37°C for 3 hours). The same number of cells was analysed. (c) Results obtained after fob1 deletion in the rpb1–1 ts strain. (d) To control the effect of the temperature on DNA replication, a wild type strain (BY4741) was used. (e) Results obtained after the isolation of DNA from a fob1Δ strain containing approximately 190 or 25 rDNA copies. A higher exposure image is shown on the right for the 25 copies strain.