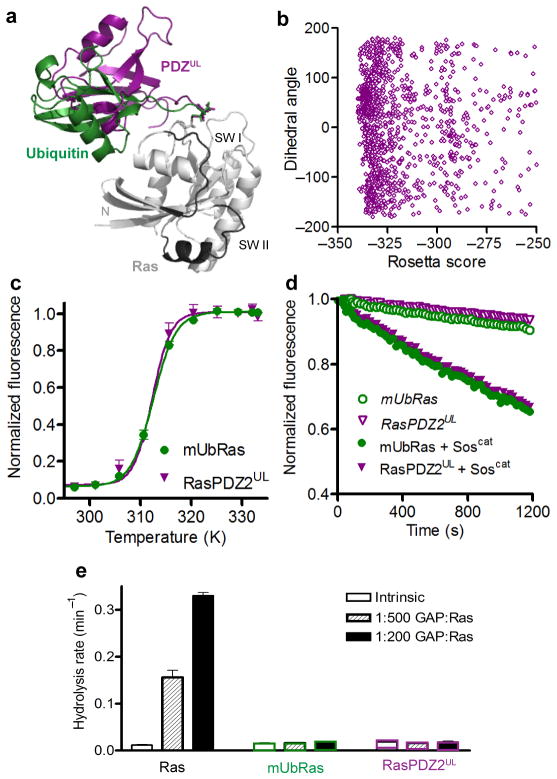
Figure 6.
Modification of Ras with PDZ2 resembles modification with Ubiquitin. (a) Rosetta model of Ras (5P21) in grey modified at position 147 with Ubiquitin (1UBQ) in green and PDZUL (3LNX) in purple. (b) The distribution of PDZUL orientations relative to Ras plotted against Rosetta energy scores for the chemical linkage. This plot follows the scheme of Figure 3B. (c) Thermal stability of Ras and RasPDZ2 with the Ubiquitin linker (RasPDZ2UL) measured by ABD–F incorporation as a function of temperature. Results are the mean ± s.d. (n=4). (d) Nucleotide dissociation reaction for RasPDZ2UL and mUbRas loaded with MANT–GDP in the absence and presence of a 1:1 molar ratio of Ras to Soscat. Results are the mean ± s.d. (n=4). (e) Single–turnover GTP hydrolysis for Ras, RasPDZ2UL, and mUbRas in the presence of GAP–334 at a molar ratio of 1:500 and 1:200 GAP:Ras. Results are the mean ± s.d. (n=6).