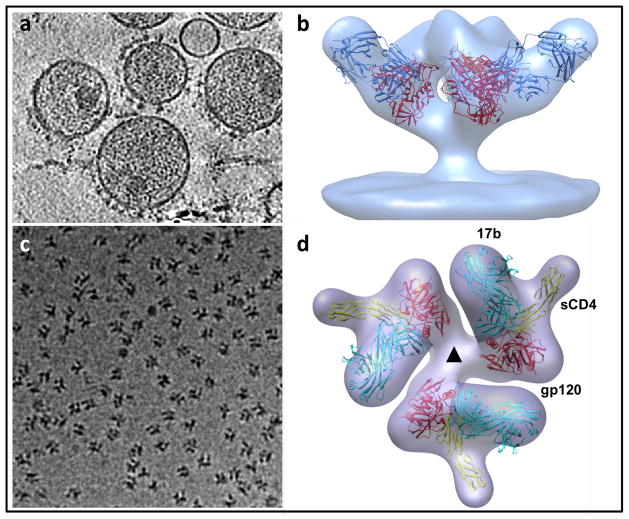
Figure 4.
Structural analysis of membrane protein complexes using cryo-electron tomography combined with sub-volume averaging. (a) Tomographic slice through a field of HIV recorded from a specimen grid that was plunge-frozen and stored at liquid nitrogen temperatures. In this image, the viral membrane is decorated with trimeric envelope glycoproteins, which are required for viral entry into target cells. (b) Density map at ~ 20 Å resolution of the trimeric envelope glycoproteins complexed with the neutralizing antibody VRC01. The map was obtained by missing wedge-corrected, subvolume averaging of cryo-electron tomographic images. The map was then fitted with three copies of the X-ray crystallographically determined structure for the complex of monomeric gp120, a portion of the HIV envelope glycoprotein, complexed with VRC01. (c) Projection image of individual molecular complexes of soluble trimeric envelope glycoproteins from human immunodeficiency virus (HIV; strain KNH1144). (d) Density map at ~ 20 Å resolution of the complex of HIV envelope glycoproteins (molecular weight of polypeptide portion ~ 240 kDa) with soluble CD4 (molecular weight ~ 24 kDa) and Fab fragment (molecular weight ~ 50 kDa). The map is fitted with three copies of the structure of the ternary complex of monomeric gp120, sCD4 and 17b Fab determined by X-ray crystallography. Figure panels adapted from[73] and [74].