Abstract
Three-dimensional (3D) reconstruction and examination of tissue at microscopic resolution have significant potential to enhance the study of both normal and disease processes, particularly those involving structural changes or those in which the spatial relationship of disease features is important. Although other methods exist for studying tissue in 3D, using conventional histopathological features has significant advantages because it allows for conventional histopathological staining and interpretation techniques. Until now, its use has not been routine in research because of the technical difficulty in constructing 3D tissue models. We describe a novel system for 3D histological reconstruction, integrating whole-slide imaging (virtual slides), image serving, registration, and visualization into one user-friendly package. It produces high-resolution 3D reconstructions with minimal user interaction and can be used in a histopathological laboratory without input from computing specialists. It uses a novel method for slice-to-slice image registration using automatic registration algorithms custom designed for both virtual slides and histopathological images. This system has been applied to >300 separate 3D volumes from eight different tissue types, using a total of 5500 virtual slides comprising 1.45 TB of primary image data. Qualitative and quantitative metrics for the accuracy of 3D reconstruction are provided, with measured registration accuracy approaching 120 μm for a 1-cm piece of tissue. Both 3D tissue volumes and generated 3D models are presented for four demonstrator cases.
The three-dimensional (3D) reconstruction and examination of tissue at microscopic resolution have significant potential to enhance the study of normal and disease processes, particularly those involving structural changes or those in which the spatial relationship of disease features is important. Its application to, or combination with, techniques, such as immunohistochemistry (IHC) or in situ hybridization, adds further value by allowing understanding of additional phenotypic or functional information. Initial applications have been concerned with investigating the anatomical features and microarchitecture of normal tissue,1 tumor invasion, growth factor expression, and localization of therapeutic targets in relation to microvasculature and studying gene expression (eg, in developing mouse embryos2 and the developing human brain3).
Competing alternative techniques to 3D tissue reconstruction using individually stained serial sections, and alternative nondestructive 3D imaging techniques, include optical projection tomography,4 3D imaging with ultrasonography,5 microscopic magnetic resonance imaging, or X-ray microcomputed tomography, confocal laser scanning or multiphoton microscopy, and serial block face imaging (eg, episcopic fluorescence image capture and high-resolution episcopic microscopy).6 Although all are mature technologies, which are used in research practice, each have limitations, whereas using conventional histopathological characteristics has significant advantages because it allows for conventional histopathological staining and interpretation techniques.
More conventional methods of 3D histopathological analysis use proved and simple laboratory techniques to study structure, function, and disease manifestations. Examples include the use of photomicrographs and customized automated desktop software,7,8 in which a digital camera mounted on a light microscope captured images from serial sections of cervical carcinoma invasion. An extension of this is the large-image microscope array,9 whereby sectioning, imaging, and reconstruction were used to reconstruct whole organs. A further example of a fully integrated system of 3D reconstruction was previously described,10 in which a tissue processing, sectioning, slide scanning, and reconstruction system was fully automated and integrated into one process. Although these are useful methods for 3D reconstructions, particularly using an integrated system, the images and 3D reconstruction are limited by low resolution.
Other limiting factors for conventional 3D histopathological analysis include the time and difficultly associated with acquiring many images with a microscope instead of an automated whole slide scanner,11–13 the absence of a fully integrated system for reconstruction,14 and the significant amount of time associated with manual input previously required to guide 3D reconstruction.13
In light of these limiting factors, we have developed novel 3D histopathological software, which uses automated virtual slide scanners to generate high-resolution digital images and produce 3D tissue reconstructions at a cellular resolution level. It can be used on any stained tissue section (eg, H&E, IHC, or special stains or chromogenic in situ hybridization). It is based on a general image-based registration algorithm, which is reasonably robust over a wide variety of data (ie, it is not tuned to a specific data type/application) and operates using an integrated system that requires minimal manual intervention once the slides are divided into sections, stained, or mounted. The virtual slide scanners digitize automatically, and the software communicates with the image-serving software, which aligns the images and produces visualization in one integrated package. It uses high-resolution registration and high-performance computing that takes advantage of parallel computing using the OpenMP library in C++ (Microsoft, Redmond, WA) to use all available cores (n = 8) of our server. It also uses a novel means of multilevel registration (described later), in which the user can manually select a region, zoom in, and reregister the area. It then uses data fusion techniques to visualize microanatomical and functional information in conjunction with the structural 3D reconstruction and novel data visualization techniques to allow researchers to explore the resulting large data sets.
In developing this novel 3D histopathological software, we are seeking to address a clinical and research need for high-resolution 3D microscopy. Many fields, including tumor biology, embryology, and transgenic models, would benefit from correlation of structure and function in 3D, but no current technology can integrate tissue microarchitecture, cellular morphological characteristics, and function on large tissue samples.
This system has been used to generate 300 separate 3D tissue volumes from eight different tissue types, using a total of 5500 virtual slides comprising 1.45 TB of primary image data. This article describes the application of this method in four case studies.
Materials and Methods
Tissue Preparation
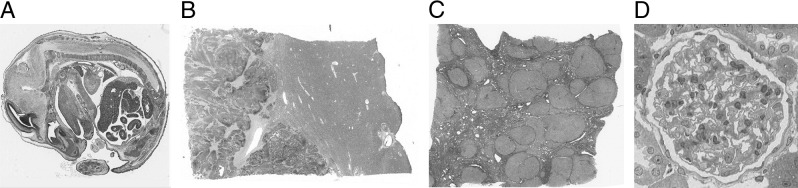
The following data sets were prepared (all shown in Figure 1): data set A, 111 sections of an 18-day post-fertilization mouse embryo (stained with H&E) (Figure 1A); data set B, 70 sections from a human liver containing a deposit of metastatic colorectal carcinoma adjacent to a blood vessel (stained with H&E) (Figure 1B); data set C, 100 sections from a cirrhotic human liver infected with hepatitis C (stained with picrosirius red) (Figure 1C); and data set D, 130 sections of a single rat glomerulus (stained with solochrome cyanin/phloxin) (Figure 1D). All human tissue was surplus surgical tissue with local ethical approval. The CD1 mouse embryo was obtained from a 10-week-old time-mated female (mated with a stud of the same strain), and the rat glomerulus was imaged from the kidney of a 12-week-old male Sprague-Dawley rat. Both were sacrificed under Schedule 1 of the Animals (Scientific Procedures) Act, 1986, under Project License PPL 40/2917.
Figure 1.
Examples of original images (axial views) from data sets: an 18-day mouse embryo stained with H&E (A), human liver containing a deposit of metastatic colorectal carcinoma stained with H&E (B), human liver with hepatitis C stained with picrosirius red (C), and a single rat glomerulus stained with solochrome cyanin/phloxin (D).
Tissue sections for data sets A, B, and C were prepared using standard histological techniques, as follows. Tissue was formalin fixed for 2 to 3 days, processed in a Leica ASP 200 tissue processor (Leica Microsystems, UK; Milton Keynes, UK) for 48 hours, and paraffin embedded. Serial sections (5 μm thick) were cut using a standard microtome, and selected sections (eg, every seventh section for data set A, giving a gap of 35 μm between images; every fifth section for data set B, giving a gap of 25 μm between images; and every 20th section for data set C, giving a gap of 100 μm between images) were stained using H&E and picrosirius red for the hepatitis C liver. Glass slides were scanned using Aperio Technologies, Inc. (San Diego, CA), T2 and T3 scanners using ×20 (for both of the human liver cases) and ×40 (for the embryo and glomerulus) objectives, producing images with a final resolution of 0.23 and 0.46 μm per pixel, respectively. Tissue sections for data set D were prepared using specialized histological techniques, as follows. Tissue was formalin fixed, processed, and embedded in LR Gold acrylic resin (Sigma-Aldrich, Poole, Dorset, UK). Serial sections (0.5 μm thick) were cut using an ultramicrotome, and every single section was selected and stained using solochrome cyanin/phloxin. The glass slides were scanned using Aperio Technologies, Inc., T2 and T3 scanners using a ×40 objective with a doubling magnifier, producing images with a final resolution of 0.92 μm per pixel. Data sets A, B, and C were then used as described in Sequential Image-Based Registration, and data set D was used as described in Interactive Multilevel Registration.
Sequential Image-Based Registration
Scanned data sets were uploaded into our customized software and registered using a sequential slice-to-slice image-based registration approach. One virtual slide (the section closest to the center of the tissue because this section generally contains the most tissue and allows for the definition of the volume size before registration commences) was used as a reference. Serial sections directly next to this reference section were nonrigidly aligned to the reference using a slice-to-slice image-based registration technique. Alignment proceeds out from the center, with subsequent images aligned to their neighbors. The set of aligned images were then concatenated to form a 3D volumetric data set. The slice-to-slice image registration approach used was a multistage method based on an extension of phase correlation.15 This method allows rotation and scale to be recovered (in addition to translation) by converting the images to log-polar coordinates. Images are first aligned rigidly (ignoring scale) by subsampling the virtual slides by a factor of 64. This result is then used as input to a nonrigid registration method that divides the image into a set of regularly spaced square patches that are individually aligned.15 A global (whole image) non–rigid B-spline–based transform is estimated using a robust least-squares error-minimizing estimation method to approximate a set of point translations derived from the set of rigid patch transforms (five points are derived from each patch, one at each corner and one at the center). This method was applied at multiple increasing image resolutions and B-spline grid sizes (zoom 1/64 B-spline 3 × 3, zoom 1/32 B-spline 4 × 4, zoom 1/16 B-spline 5 × 5, and zoom 1/8 B-spline 6 × 6), with the result of the previous application initializing the next application. This was all integrated into a user-friendly software package that obtains images from the image server and generates visualizations of the images with a few button clicks. Qualitative analysis was performed on all data sets to determine the accuracy of each reconstruction; in addition, quantitative analysis was performed on test data sets (see Results). On completion of the rigid and nonrigid transform, a volume was generated for each data set, in which the axial, coronal, and sagittal views could be studied.
Interactive Multilevel Registration
The image shown in Figure 1D has a width/height approximately 512 times smaller than the current maximum resolution of slide scanners. Thus, small errors in calculated low-resolution transforms are scaled up to become large errors at the native resolution. The worst-case registration accuracy when registering entire virtual slides can be as large as a single cell (described later). This is the result of a B-spline with many knots being required to represent subcellular-level deformations over the entire image (which is difficult to estimate robustly in the presence of local tissue and imaging irregularities). To obtain subcellular accuracy reconstructions, we have implemented an interface, which is integrated into our software, by which the user can interactively select subareas of the image to register at higher resolution, using the lower-resolution output as an initial solution (ie, using a B-spline with relatively few knots). This is illustrated in Figure 2. This method is similar in concept to the multilevel resolution registration method previously detailed,16 but uses multiple resolution levels for nonrigid registration as opposed to using a single high-resolution level with point features for nonrigid registration. The output from the whole image registration is used as an initial solution for this registration, which is based on the method previously described.17 To achieve this subcellular-level accuracy, thin sections in which the same cells are visible in successive sections are required.
Figure 2.
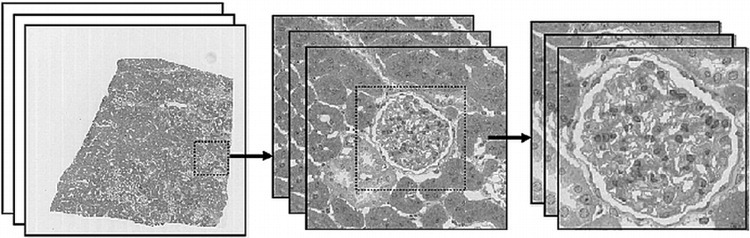
Performing multilevel registration at successive image magnifications using interactive subimage selection.
3D Rendering, Segmentation, and Visualization
The volume generated is then used to generate a 3D volume rendering and segmentation of each data set. This software is based on software ray tracing and code from the Visualization Toolkit (Kitware, Clifton Park, NY; available at http://www.vtk.org),18 which supports 3D volume rendering of raw data in color and isosurfacing of segmentations; both are integrated into the reconstruction software. The 3D volume rendering software has clip planes controlled by sliders, in conjunction with a zooming functionality, by which the user can manipulate the x, y, and z axes to zoom in on and view areas of interest.
The interactive segmentation process involves users going through each image individually within the data set and manually segmenting (highlighting) the area they want to visualize. The software we have developed facilitates interactive segmentation of 3D structures using a variety of methods (including purely manual annotation). Of particular utility is a method we call color exemplar thresholding, in which the user selects an example color by clicking with the mouse. All pixels with an RGB color within a user-specified threshold of this example color are annotated. Alternatively, this approach may be used within a region-growing framework19 to segment only spatially connected similar pixels, using the selected location as a seed and color exemplar thresholding as the uniformity predicate. After segmentation is complete, the user can then perform isosurfacing of the segmentation using marching cubes20 and mesh decimation (using a code from the Visualization Toolkit18) to generate a 3D color visualization. Isosurfacing of segmentations can be performed on numerous structures/components of the data set, as demonstrated in Results, and can be used to selectively volume render and view different parts of the reconstructed volumes independently of one another with arbitrary topological features (in addition to more conventional cut planes).
Qualitative and Quantitative Performance
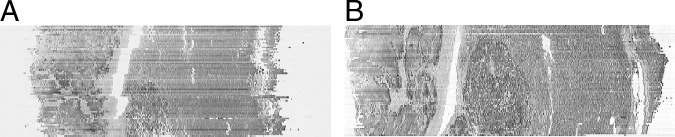
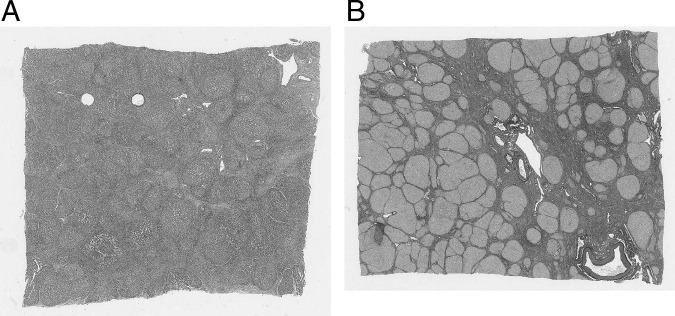
Qualitative performance for all data sets was judged by viewing the reconstructed volumes in coronal (Figure 3) and sagittal views; these views contain one row from each image in the set. Misalignment errors can be seen as discontinuities between rows. This was typically performed after the completion of rigid registration. In the event that there was a registration failure (Figure 3A), the problem images were identified and translated and/or rotated manually using a manual correction program, which is integrated into our software. This ensured that when the manually corrected images were rigidly registered again, and subsequently nonrigidly registered, they were successfully aligned (Figure 3B). Quantitative performance was performed on two test data sets (Figure 4) containing roughly similar features (eg, holes and blood vessels aligned perpendicular to the sectioning direction), and these features were annotated using Aperio ImageScope software version 10 (Aperio Technologies, Inc.). Annotations (consisting of closely spaced point sets) were aligned using the registration transform computed using our software, and the difference between aligned annotations was quantified using the standard Hausdorff distance measure (ie, maximum distance).21 Distances were quantified, as shown in Figure 4. Thirty-nine and thirty-one successive serial sections were used, repectively, to measure section-to-section accuracy for all stages of image registration and to measure accumulated error between images 10 sections apart. The accuracy of this evaluation is limited by the accuracy of the manual annotations and the variation in shape from one section to another (which we estimate to be in the order of 50 to 100 μm).
Figure 3.
A: Coronal view through the center of an image stack (Figure 1A) showing failures in rigid registration. B: Coronal view through the center of the same image stack (data set B, liver with metastatic colorectal carcinoma) showing reconstruction has worked successfully after subsequent manual correction and nonrigid registration.
Figure 4.
A: Axial view of cirrhotic liver tissue stained with H&E with highlighted synthetic holes used for registration accuracy metrics. B: Axial view of cirrhotic liver tissue stained with picrosirius red with a central single blood vessel used for registration accuracy metrics.
Results
Data set volume and segmentation sizes were as follows: the mouse embryo (data set A: 244-MB volume, 82-MB segmentation), the human liver tissue with metastatic colorectal carcinoma (data set B: 315-MB volume, 95-MB segmentation), the human liver cirrhotic tissue with hepatitis C (data set C: 347-MB volume, 110-MB segmentation), and the rat glomerulus (data set D: 665-MB volume, 221-MB segmentation). The time it takes for an entire reconstruction, from fixation at the start to segmentation at the end, for each case is shown in Table 1. Most of the time taken is computational time to scan and reconstruct the images; of the human time required, most is for sectioning and staining. Only a small proportion of technician time is spent performing and quality controlling the 3D reconstruction (ie, checking if rigid registration has worked successfully and altering the zoom and B-spline before initiating the next application level).
Table 1.
Approximate Time Taken for Each 3D Reconstruction Case
| Variable | Mouse embryo | Human liver carcinoma | Human hepatitis C liver | Single rat glomerulus |
|---|---|---|---|---|
| Time taken by human | ||||
| Sectioning time (approximately 50 sections/hour) (minutes) | 130 | 100 | 120 | 160 |
| Staining time (minutes) | 80 | 80 | 160 | 40 |
| Segmentation (optional) (minutes) | 180 | 120 | 180 | 180 |
| Total human time (minutes) | 390 (6.5 hours) | 300 (5 hours) | 460 (7.7 hours) | 380 (6.3 hours) |
| Computational time⁎ | ||||
| Slide scanning (1 image/slide apart from the rat glomerulus: 18 images/slide; 8 slides in total) | 3330 | 350 | 500 | 1080 |
| Rigid and nonrigid registration | 720 | 540 | 660 | 960 |
| Total computer processing time (minutes) | 4050 (67.5 hours) | 890 (14.8 hours) | 1160 (19.3 hours) | 2040 (34 hours) |
Computational processing is unsupervised by humans.
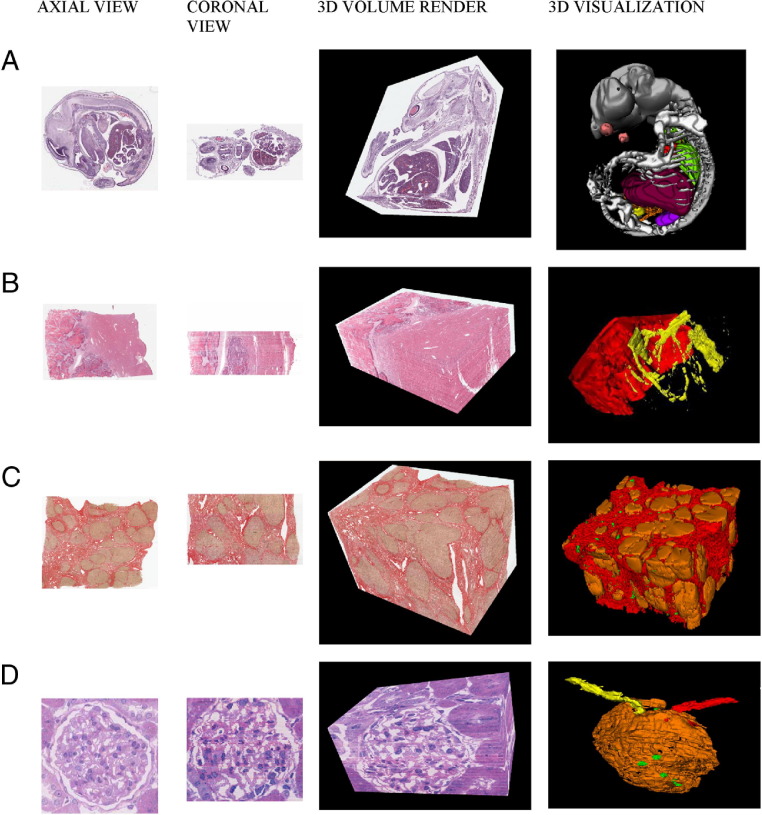
The 3D reconstruction results for all three data sets are shown in Figure 5. The 3D volume rendering of Figure 5A demonstrates the mouse brain, eye, gut, and liver, providing information on the anatomical morphological and morphometric features of different components. The 3D visualization demonstrates the anatomical features of the mouse embryo, including the brain and spinal cord, liver, lungs, heart, kidneys, bladder, gut, eyes, and skeletal formation. Possible applications include providing anatomical and expression data and virtual archiving of transgenic models. The 3D volume rendering of Figure 5B shows tumor morphological features and vessels running adjacent to and into the metastatic adenocarcinoma; this rendition could provide insight into tumor vasculature and its response to anti-angiogenic agents and the development of such lesions. The 3D visualization demonstrates bifurcation of the vessels in the liver tissue (yellow) with an enveloping metastatic colorectal carcinoma front (red). The 3D volume rendering of Figure 5C provides information on nodular shape and relationship to cirrhotic fibrosis. It also identifies the vessels running throughout the tissue, thus providing possible insight into vascular and ductular structure in relation to fibrogenesis and hepatic stellate cell and myofibroblast activity. The 3D visualization shows cirrhotic nodules (orange) surrounded by fibrosis (scarring, red), and the vasculature (green) can also be seen. It demonstrates the software's potential for provision of information on the development of the disease and may refine diagnosis. It also demonstrates the software's utility on a specialized stain (picrosirius red). The 3D volume rendering of Figure 5D shows the single rat glomerulus surrounded by tubular basement membranes and the afferent and efferent arterioles on either side of the glomerulus. It demonstrates the intricate nature of the glomerulus and provides information on cellular components, such as the capillaries and mesangial cells, within the Bowman's capsule. The 3D visualization shows the Bowman's capsule and the proximal convoluted tubule (orange), the afferent arteriole (red), the efferent arteriole (yellow), and selected individual mesangial cell nuclei (green). It also demonstrates the software's utility at a high resolution. Users can use the x, y, and z slider functions to manipulate the areas of interest they want to view in the 3D volume rendition, and different components can be turned on or off so that each can be individually viewed in the 3D visualization.
Figure 5.
Axial, coronal, and sagittal views with a 3D volume rendition and 3D visualization of anatomical features of the mouse embryo (A), metastatic colorectal carcinoma in human liver tissue (B), cirrhotic human liver tissue infected with hepatitis C (C), and a single rat glomerulus (D). In B, the tumor is marked in red and the blood vessels in the adjacent liver tissue are marked in yellow. Colors in the first three columns are original colors of the stained tissue. Colors in the last column are pseudocolors chosen to highlight differences between tissue types.
Qualitative performance results are demonstrated in Figure 3, which shows two different coronal views through the center of two separate reconstructions of data set A (both have been subject to the same registration process, detailed in the second part of Materials and Methods). Figure 3A shows at least three failure registrations, particularly the discontinuity in the blood vessel (vertical white structure), whereas Figure 3B shows no major registration failures and the blood vessel is a smoother structure. Quantitative performance (Figure 4) produced registration accuracy metrics that can be seen in Tables 2 and 3. Table 2, the accuracy metrics for the H&E case (Figure 4A), shows an approximate 75-μm average maximum error with an SD of 30 μm. In Table 3, the accuracy metrics for the picrosirius red case (Figure 4B) show an approximate 54-μm average maximum error with a standard deviation of 37 μm. The worst-case registration accuracy, when registering whole virtual slides, is approximately 1 cell so the 75- or 54-μm average maximum error is acceptable. Overall, registration accuracy over 10 sections approaches 120 μm for a piece of tissue 10,000 μm large (ie, approximately 1.2% error).
Table 2.
Liver Cirrhosis Stained with H&E Registration Accuracy Metrics Showing Typical Slice-to-Slice Registration Accuracy and Accumulated Error over 10 Sections for Each Registration Application Level
| Variable | Unregistered | Rigid | z64 (3 × 3) | z32 (4 × 4) | z16 (5 × 5) | z8 (6 × 6) |
|---|---|---|---|---|---|---|
| Typical slice: slice registration accuracy (μm) | ||||||
| Mean | 771.75 | 91.88 | 51.43 | 54.64 | 50.54 | 49.17 |
| SD | 429.77 | 50.88 | 32.13 | 32.75 | 34.44 | 31.83 |
| Accumulated error over 10 sections (μm) | ||||||
| Mean | 915.81 | 224.38 | 140.67 | 110.94 | 113.18 | 112.74 |
| SD | 497.28 | 100.59 | 73.19 | 67.68 | 75.83 | 70.67 |
Data are given as the Hausdorff distance.
Table 3.
Liver Cirrhosis Stained with Picosirius Red Registration Accuracy Metrics Showing Typical Slice-to-Slice Registration Accuracy and Accumulated Error over 10 Sections for Each Registration Application Level
| Variable | Unregistered | Rigid | z64 (3 × 3) | z32 (4 × 4) | z16 (5 × 5) | z8 (6 × 6) |
|---|---|---|---|---|---|---|
| Typical slice-to-slice registration accuracy (μm) | ||||||
| Mean | 776.17 | 104.64 | 58.93 | 61.15 | 58.09 | 54.37 |
| SD | 424.36 | 77.85 | 32.68 | 37.11 | 42.52 | 37.17 |
| Accumulated error over 10 sections (μm) | ||||||
| Mean | 1013.43 | 227.92 | 141.73 | 118.44 | 130.99 | 120.36 |
| SD | 462.08 | 108.37 | 61.92 | 53.98 | 81.38 | 88.32 |
Measurements are calculated as the Hausdorff distance in micrometers (µm).
Discussion
We report a novel means of 3D histological reconstruction that integrates whole slide scanning, image serving, registration, and visualization into one integrated user-friendly package. It produces high-resolution 3D reconstructions with minimal user interaction subsequent to sectioning, staining, and mounting (these processes can be automated,22 but we did not have access to such equipment). We have also demonstrated that this software is accurate and robust because we have achieved qualitatively similar results from a range of different tissues, embedding autostains and histochemical stains, using serial and step sections. We propose that this system provides the opportunity for increasing the use of 3D histopathological analysis as a routine research tool.
There is a range of uses for this software. First, availability of easily produced 3D reconstructions and 3D visualizations enable the exploration of large 3D data sets, which allows for the extensive analysis of tissue microanatomical and morphological structure. Second, automated or semiautomated measurement of 3D volumes can be performed (eg, to quantify vascular volume or structure). Third, automating histochemical and IHC staining in a 3D volume provides the ability to study structural and functional information simultaneously (eg, to quantify protein expression at the invasive front of a tumor or organ-specific growth factor expression in developing embryos/placentas). Finally, it can assist the development of novel in vivo microscopic imaging. Imaging modalities, such as in vivo microscopy (eg, of dysplasia in gastrointestinal endoscopy) and high-resolution radiological imaging, are being developed. The availability of robust 3D tissue visualization will allow the validation of these novel imaging techniques against the gold standard, and this could enhance pathological-radiological-clinical correlation.
Other experimenters have used 3D reconstruction to similar ends (reconstruction of colorectal carcinoma liver metastases23 or the invasive front of cervical carcinoma7,8), but such studies are limited in size and scope by the need to acquire photomicrographs, the lack of available and easily usable software, and the unavailability of high-resolution images that can be iteratively reconstructed at any scale. Although others12 have also used virtual slides to improve the speed of image acquisition, iterative semiautomatic registration using the high-resolution whole slide image has not yet been performed. Alternative techniques of 3D reconstruction have also been developed but are limited. Optical projection tomography4 offers good reconstruction while keeping the specimens intact but is limited in resolution and to small sample sizes. 3D imaging with ultrasonography,5 magnetic resonance imaging, or X-ray microcomputed tomography is nondestructive but is also limited by the maximum resolution obtainable and a lack of simultaneous functional information. Confocal laser scanning microscopy and multiphoton microscopy offer high-resolution 3D reconstruction, but sample sizes are small so architectural context may be lost. Serial block face imaging (episcopic fluorescence image capture and high-resolution episcopic microscopy)6 provides high resolution over hundreds of micrometers but is limited because it is not possible to repeatedly image a region after the block has been cut, and IHC staining can only be performed before tissue embedding.
Future studies may use specialist equipment, as previously detailed,22 which will address the major bottleneck in our 3D reconstruction process, which is the time taken to prepare serial or step sections on glass slides. Also, the use of a fully automated staining machine will further minimize the amount of manual input in this process.
Future development efforts will focus on developing whole slide 3D visualization at full resolution. Although alignment and reconstruction may be performed at the full (native) resolution of the slides, 3D visualization of the full reconstruction is not possible at this resolution because of memory and screen resolution constraints (eg, 3D visualizations obtained from this study were limited to 2000 × 2000 pixel images of >100 sections); however, subvolumes may be visualized. In future work, we plan to develop a system for intuitive navigation of such data sets at all resolutions. This will produce, in effect, a 3D microscope for gigapixel-sized 3D volumes that would require high-resolution displays, such as a Powerwall,24 for viewing.
Acknowledgments
We thank Mike Hale, Martin Waterhouse, David Turner, and Alex Wright (Leeds Institute of Molecular Medicine Digital Pathology Group) for scanning and computing assistance.
Footnotes
Supported by funding from the National Cancer Research Institute informatics initiative, Leeds Teaching Hospital Trust Research and Development, National Institute for Health Research, West Yorkshire Comprehensive Local Research Network, and UK Department of Health.
This work was partially funded through WELMEC, a Centre of Excellence in Medical Engineering funded by the Wellcome Trust and EPSRC (WT 088908/Z/09/Z).
The views expressed in this article are those of the authors and not those of the funding bodies.
References
- 1.Kaufman M.H., Brune R.M., Baldock R.A., Bard J.B.L., Davidson D. Computer-aided 3D reconstruction of serially sectioned mouse embryos: its use in integrating anatomical organization. Int J Dev Biol. 1997;41:223–233. [PubMed] [Google Scholar]
- 2.Han L., van Hemert J.I., Baldock R.A. Automatically identifying and annotating mouse embryo gene expression patterns. Bioinformatics. 2011;27:1101–1107. doi: 10.1093/bioinformatics/btr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X., Lindsay S., Baldock R. From spatial-data to 3D models of the developing human brain. Methods. 2010;50:96–104. doi: 10.1016/j.ymeth.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Quintana L., Sharpe J. Optical projection tomography of vertebrate embryo development. Cold Spring Harb Protoc. 2011;2011:586–594. doi: 10.1101/pdb.top116. [DOI] [PubMed] [Google Scholar]
- 5.Prager R.W., Ijaz U.Z., Gee A.H., Treece G.M. Three-dimensional ultrasound imaging. Proc Inst Mech Eng H. 2010;224:193–223. doi: 10.1243/09544119JEIM586. [DOI] [PubMed] [Google Scholar]
- 6.Denk W., Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wentzensen N., Braumann U.D., Einenkel J., Horn L.C., von Knebel Doeberitz M., Löffler M., Kuska J.P. Combined serial section-based 3D reconstruction of cervical carcinoma invasion using H&E/p16INK4a/CD3 alternate staining. Cytometry A. 2007;71:327–333. doi: 10.1002/cyto.a.20385. [DOI] [PubMed] [Google Scholar]
- 8.Braumann U.D., Kuska J.P., Einenkel J., Horn L.C., Loffler M., Hockel M. Three-dimensional reconstruction and quantification of cervical carcinoma invasion fronts from histological serial sections. IEEE Trans Med Imaging. 2005;24:1286–1307. doi: 10.1109/42.929614. [DOI] [PubMed] [Google Scholar]
- 9.de Ryk J., Namati E., Reinhardt J.M., Piker C., Xu Y., Liu L., Hoffman E.A., McLennan G. A whole organ serial sectioning and imaging system for correlation of pathology to computer tomography. Pro Biomed Opt Imag. 2004;5:224–234. [Google Scholar]
- 10.Onozato ML, Merren M, Yagi Y: Automated 3D-reconstruction of histological sections. Presented at the International Academy of Digital Pathology Conference, Quebec, Canada, 2011
- 11.Al-Janabi S., Huisman A., Van Diest P.J. Digital pathology: current status and future perspectives. Histopathology. 2011 doi: 10.1111/j.1365-2559.2011.03814.x. [DOI] [PubMed] [Google Scholar]
- 12.Wu M.L.C., Varga V.S., Kamaras V., Ficsor L., Tagscherer A., Tulassay Z., Molnar B. Three-dimensional virtual microscopy of colorectal biopsies. Arch Pathol Lab Med. 2005;129:507–510. doi: 10.5858/2005-129-507-TVMOCB. [DOI] [PubMed] [Google Scholar]
- 13.Petrie I.A., Flynn A.A., Pedley R.B., Green A.J., El-Emir E., Dearling J.L., Boxer G.M., Boden R., Begent R.H. Spatial accuracy of 3D reconstructed radioluminographs of serial tissue sections and resultant absorbed dose estimates. Phys Med Biol. 2002;47:3651–3661. doi: 10.1088/0031-9155/47/20/307. [DOI] [PubMed] [Google Scholar]
- 14.Namati E., De Ryk J., Thiesse J., Towfic Z., Hoffman E., McLennan G. Large image microscope array for the compilation of multimodality whole organ image databases. Anat Rec (Hoboken) 2007;290:1377–1387. doi: 10.1002/ar.20600. [DOI] [PubMed] [Google Scholar]
- 15.De Castro E., Morandi C. Registration of translated and rotated images using finite fourier transforms. IEEE Trans Pattern Anal Mach Intell. 1987;9:700–703. doi: 10.1109/tpami.1987.4767966. [DOI] [PubMed] [Google Scholar]
- 16.Kun H, Lee C, Ashish S, Tony P: Fast automatic registration algorithm for large microscopy images. Presented at the Life Science and Applications Workshop, Bethesda, MD, 2006
- 17.Magee D., Treanor D., Quirke P. 2008. A new image registration algorithm with application to 3D histopathology: Microscopic Image Analysis with Applications in Biology. New York, NY. [Google Scholar]
- 18.Schroeder W., Martin K., Lorensen W.E. ed 3. Kitware Inc.; New York: 2003. The Visualization Toolkit: An Object-Oriented Approach to 3D Graphics. [Google Scholar]
- 19.Efford N. Addison-Wesley; Harlow, England, and New York: 2000. Digital Image Processing: A Practical Introduction Using Java; p. 340. [Google Scholar]
- 20.Lorensen W.E., Cline H.E. Marching cubes: a high resolution 3D surface construction algorithm. SIGGRAPH Comput Graph. 1987;21:163–169. [Google Scholar]
- 21.Huttenlocher D.P., Klanderman G.A., Rucklidge W.J. Comparing images using the Hausdorff distance. IEEE T Pattern Anal. 1993;15:850–863. [Google Scholar]
- 22.Mitsunori K T.H., Ken'ichi K., Yoshimitsu F., Akihiro O., Hiroshi N., Hisashi I. Development of the automatic thin sectioning microtome system for light microscopy: the machine to mount sections on the object glass automatically by using static electricity. J Jpn Soc Precision Eng. 2002;68:1605–1610. [Google Scholar]
- 23.Griffini P., Smorenburg S.M., Verbeek F.J., van Noorden C.J. Three-dimensional reconstruction of colon carcinoma metastases in liver. J Microsc. 1997;187:12–21. doi: 10.1046/j.1365-2818.1997.2140770.x. [DOI] [PubMed] [Google Scholar]
- 24.Treanor D., Jordan-Owers N., Hodrien J., Wood J., Quirke P., Ruddle R.A. Virtual reality Powerwall versus conventional microscope for viewing pathology slides: an experimental comparison. Histopathology. 2009;55:294–300. doi: 10.1111/j.1365-2559.2009.03389.x. [DOI] [PubMed] [Google Scholar]