Abstract
Affinity maturation refines a naive B-cell response by selecting mutations in antibody variable domains that enhance antigen binding. We describe a B-cell lineage expressing broadly neutralizing influenza virus antibodies derived from a subject immunized with the 2007 trivalent vaccine. The lineage comprises three mature antibodies, the unmutated common ancestor, and a common intermediate. Their heavy-chain complementarity determining region inserts into the conserved receptor-binding pocket of influenza HA. We show by analysis of structures, binding kinetics and long time-scale molecular dynamics simulations that antibody evolution in this lineage has rigidified the initially flexible heavy-chain complementarity determining region by two nearly independent pathways and that this preconfiguration accounts for most of the affinity gain. The results advance our understanding of strategies for developing more broadly effective influenza vaccines.
Keywords: immunity, antigen recognition, X-ray crystallography
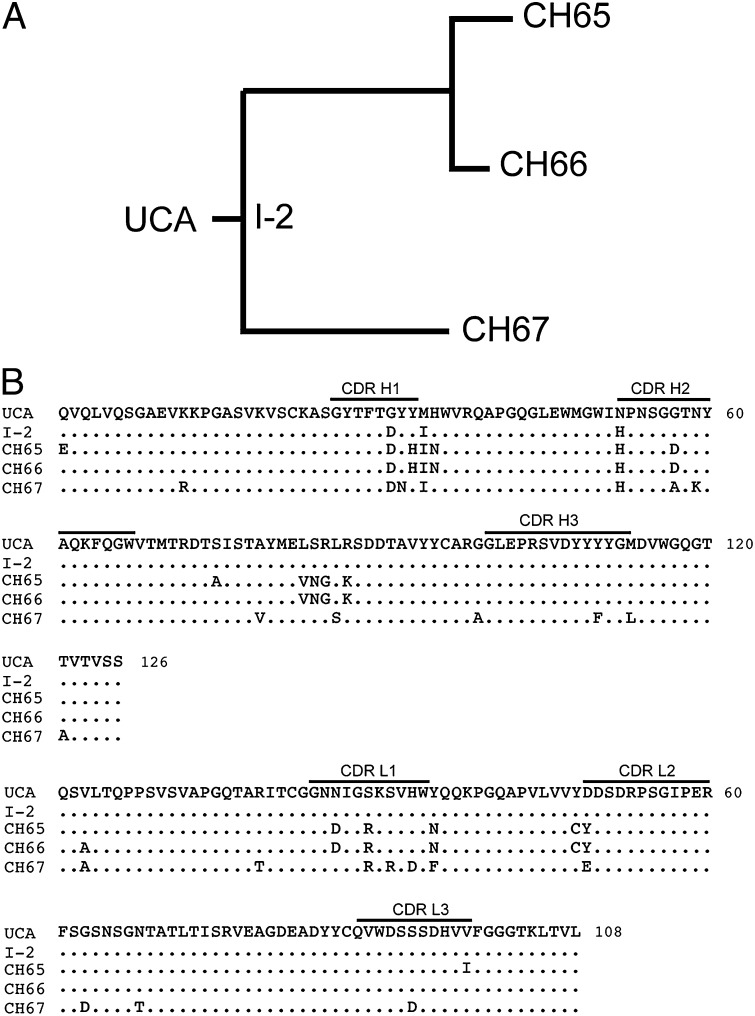
Exposure to a novel antigen, whether by infection or vaccination, induces an initial naive B-cell response. Cells bearing B-cell receptors (BCRs) that bind the antigen in question, even with relatively low affinity, proliferate selectively. In the continued presence of antigen, additional proliferation, accompanied by somatic hypermutation of the rearranged Ig heavy- and light-chain genes, leads to selection of cells with BCRs (and secreted antibodies) that bind more tightly to antigen than their precursors—a process known as affinity maturation (1–3). Recent methodological advances make it possible to study the history of this process in a given subject by isolating a number of individual B cells at a suitable time point after vaccination or infection and cloning their recombined heavy- and light-chain variable regions (4–6). If a subset of the variable regions thus identified derives from the same progenitor, one can infer the clonal lineage that gave rise to the observed genes, including the unmutated common ancestor (UCA) and the other unobserved intermediates at the interior nodes of the clonal tree with tips that are the genes of the mature antibodies (Fig. 1A). Structural and biochemical changes that occur during affinity maturation can be analyzed, and the mechanism of affinity enhancement elucidated.
Fig. 1.
Inferred CH65–CH67 lineage and sequences. (A) Members of the lineage include the UCA, I-2, and three mature B-cell clones, CH65, CH66, and CH67. (B) Sequence alignment of the heavy- and light-chain variable domains. CDR1, CDR2, and CDR3 loops for each chain are marked. Conservation within the lineage based on the UCA sequence is shown by a period.
The influenza B-cell clonal lineage shown in Fig. 1A derives from plasmablasts sorted from a sample taken from an adult subject 1 wk after administration of the 2007 trivalent inactivated influenza virus vaccine. It includes just three mature B-cell clones. We have shown that one member of this lineage (CH65) bears a heavy-chain complementary determining region 3 (CDR H3) loop that inserts into the HA receptor-binding pocket, mimics the influenza virus receptor sialic acid, and has unusual breadth of neutralizing capacity (31 of 36 H1 strains tested) (7). We have now extended the structural and functional analysis to the entire lineage. By determining the structure and binding properties of the UCA and intermediate 2 (I-2) Fab and comparing them with the corresponding properties of free and bound CH67 and bound CH65, we show that antibody evolution in this lineage has preconfigured the flexible CDR H3, yielding a 30- to 40-fold increase in the association rate; there is also a smaller decrease in the dissociation rate. Long time-scale molecular dynamics (MD) simulations likewise indicate that the UCA CDR H3 loop predominately assumes a range of conformations that are incompatible with binding to the HA receptor-binding pocket, whereas the CH65 and CH67 CDR H3 loops spend a substantial fraction of the simulation time in conformations close to the ones that they adopt in complex with HA.
Results
There are two distinct branches to the lineage that we have analyzed, with the nearly identical CH65 and CH66 antibodies in one branch and the quite distinct CH67 in the other branch. The branches diverge at intermediate I-2, which differs from the UCA at only three positions (Fig. 1B). Like CH65, CH67 neutralizes a large fraction of the H1 strains that we have examined (Table 1). There are a few influenza strains that are sensitive to one antibody and resistant to the other antibody, with some of the discordant strains neutralized by one and some neutralized by the other; for a number of strains, the potencies are the same. We restricted the analysis of CH67 to those strains for which we had sequence information (Fig. S1)—22 strains in all. We did not detect any obvious correlation between HA sequence and neutralization by one antibody or the other. The insertion of lysine at position 133a does not correlate with resistance, disproving a suggestion that we made in our previous paper based on a more limited set of HA sequences (7). In particular, CH67 neutralizes (with modest potency) the 2009 pandemic swine origin virus, A/California/07/2009.
Table 1.
Comparison of A/H1N1 influenza strain neutralization by human mAbs CH65 and CH67
| H1N1 virus strain | mAb minimum effective concentration (μg/mL) |
|
| CH65 | CH67 | |
| A/USSR/90/1977* | 100 | 25 |
| A/Kawasaki/6/1986 | 0.098 | 0.39 |
| A/Texas/36/1991* | Neg | Neg |
| A/Wellington/47/1992 | Neg | Neg |
| A/Florida/2/1993 | 0.012 | 0.012 |
| A/Beijing/262/1995* | 0.098 | 0.098 |
| A/Shengzhen/227/1995 | 0.012 | 0.024 |
| A/Shanghai/8/1996 | Neg | Neg |
| A/Johannesburg/159/1997 | 0.098 | 0.39 |
| A/Shanghai/2/1997 | 0.195 | 0.195 |
| A/Moscow/13/1998 | 0.012 | 0.012 |
| A/Ostrava/801/1998 | 12.5 | Neg |
| A/New Caledonia/22/1999* | 0.391 | 0.195 |
| A/Bangkok/163/2000 | 0.195 | 0.098 |
| A/Fujian/156/2000 | 0.488 | 0.049 |
| A/Chile/8885/2001 | 0.195 | 0.098 |
| A/Auckland/65/2001 | 0.195 | 0.098 |
| A/Neimenggu/52/2002† | 0.098 | 0.098 |
| A/Brazil/1403/2003 | 3.125 | 0.195 |
| A/Canada/59/2004 | 0.098 | 0.098 |
| A/Solomon Islands/03/2006*‡ | 0.024 | 0.098 |
| A/Brisbane/59/2007* | 0.098 | 0.98 |
| A/California/07/2009(swine)* | Neg | 6.25 |
*Strains included in seasonal vaccines.
†Originally reported as insensitive to mAb CH65.
‡H1 component of the vaccine received by the subject.
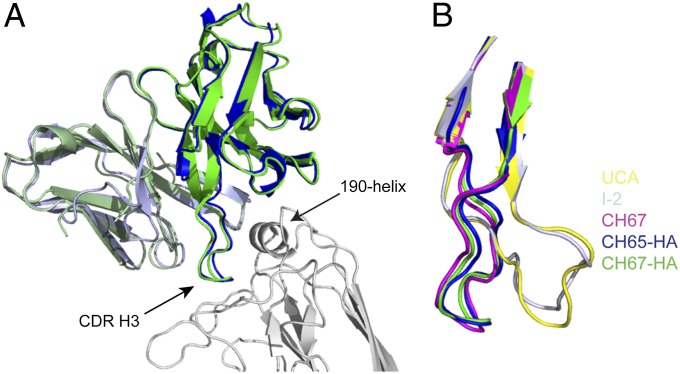
Fig. 2 shows the structures of Fab variable modules from CH65 and CH67 Abs bound with the HA head from the A/Solomon Islands/3/2006 virus—a component of the 2007 trivalent inactivated influenza virus vaccine—and compares them with the structures of the corresponding modules of the free CH67, I-2, and UCA Fabs. All of the critical interactions that we described for the CH65 complex are present in the CH67 complex, and the CDR H3 conformations of the mature Abs in complex are essentially identical to both each other and the conformation of the CDR H3 on the unbound CH67 Fab. In the latter case, there are six copies of the Fab per asymmetric unit, and all have the same CDR H3 conformation (Fig. S2). The UCA antibody binds too weakly for cocrystallization with HA head, but the crystal structure of its free Fab shows that the CDR H3, which has the same sequence as it does in CH65 and CH66 and differs from the CH67 CDR H3 by only two conservative substitutions at positions facing away from the antigen, is disordered on one of two copies in the asymmetric unit and constrained by crystal contacts to adopt a conformation on the second copy quite different from the one that it has in the other structures. Related crystal contacts cause the CDR H3 of the I-2 Fab to adopt the same constrained conformation. (Data collection and refinement statistics are in Table S1.)
Fig. 2.
Crystal structures of free and bound Fabs in the CH65–CH67 lineage. (A) CH65 and CH67 Fabs bound with HA. CH67 Fab (heavy and light chains in green and pale green, respectively), CH65 Fab (heavy and light chains in blue and light blue, respectively, from Protein Data Bank ID code 3SM5), and HA head (silver). The CDR H3 of the Fabs and the 190-helix of HA are marked for reference. (B) Comparison of free and bound Fabs. Alternative conformations of the CDR H3 loop (residues 96–117 are displayed) from the UCA (yellow) and I-2 (silver; constrained by a crystal contact), free CH67 (magenta), bound CH65 (blue), and bound CH67 (green) are displayed. All images created in PyMol.
The Fabs of CH65 and CH67 bind HA with similar association rates and affinities (Table 2). Their association rate constants are within an order of magnitude of the upper limit observed in practice for interaction of two 40- to 50-kDa globular proteins. The UCA and I-2 Fabs bind weakly, with a KD about 200-fold greater than the KD value of the mature Fabs. A slower association rate accounts for most of the difference (about two-thirds of the logarithm of the KD ratio). We can explain this observation by the disorder-to-order transition in the flexible CDR H3 loop that must occur for the UCA or I-2 to bind HA; stabilization of that loop on the mature antibodies in its bound conformation increases the likelihood of a productive association.
Table 2.
Summary of analysis of surface plasmon resonance of Fab binding to immobilized HA (A/Solomon Islands/03/2006) head
| ka′ | kd* | KD | |
| Fab | M−1s−1 | s−1 | μM |
| UCA | 4.4 ± 0.3 × 103 | 0.51 ± 0.03 | 118 ± 14 |
| I-2 | 4.0 ± 0.5 × 103 | 0.56 ± 0.01 | 142 ± 15 |
| CH65 | 1.33 ± 0.05 × 105 | 0.064 ± 0.011 | 0.49 ± 0.10 |
| CH67 | 2.37 ± 0.14 × 105 | 0.086 ± 0.012 | 0.36 ± 0.04 |
Surface plasmon resonance data were analyzed as described in Materials and Methods. The averages and SEs are for three runs each, except for CH67 (n = 6). The KD values are derived by averaging the individual KD estimates (not from the ratio of the average rates). These measurements are all for monovalent Fabs; the divalent intact antibodies bind considerably more tightly (KD between 1 and 10 nM).
MD simulations, carried out with Anton (8), a special purpose supercomputer, extend the picture of antibody binding suggested by our structural and kinetic data. We simulated the variable regions of the heavy and light chains (heavy-chain residues 1–126 and light-chain residues 1–111) and the head region (residues 51–275) of HA. All simulations were performed at a temperature of 310 K and a pressure of 1 atm.
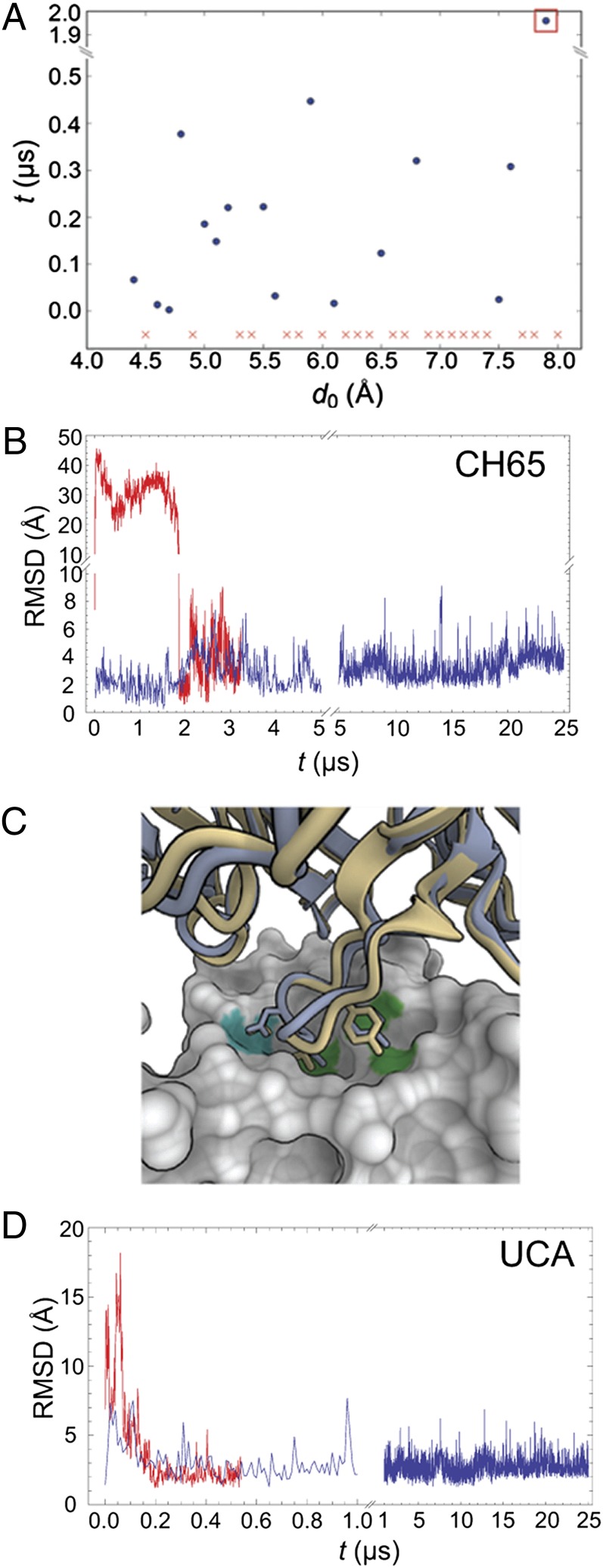
We first tested whether MD simulations can reproduce the observed antibody–HA complex structure by performing a set of simulations, in which the initial positions of HA and CH65 differed to various extents from their relative positions within the crystallographically determined CH65–HA complex (Protein Data Bank ID code 3SM5). In 19 of 37 simulations, the antibody bound to HA in a conformation similar to the one seen by X-ray crystallography (backbone rmsd < 3 Å); in 16 of these simulations, the antibody returned to its bound conformation with a backbone rmsd below 1.5 Å. Fig. 3A shows the time required for the antibody to return spontaneously to its bound conformation from various initial displacements. Fig. 3B shows the time trace of the rmsd of the antibody with respect to its crystallographic bound position in a simulation with an initial displacement of 7.9 Å, in which the antibody underwent large changes in orientation with respect to HA before settling into the correct binding position (Movie S1). These results suggest that our simulations provide a reasonable structural description of antibody–HA binding.
Fig. 3.
Long time-scale MD simulations. (A) Time required for the CH65 Fab to bind spontaneously to HA starting from various initial separations. Each of these simulations was initiated with the CH65 Fab placed at a different distance, d0, from its position within the CH65–HA complex. Details of these simulations are described in SI Materials and Methods. Each simulation was 0.5 μs in length, except for those simulations with d0 > 7.5 Å, which were 5 μs long. To determine whether the Fab had bound to HA, we aligned HA to the CH65–HA crystal structure, computed the Fab backbone rmsd with respect to its position in the complex, and considered the Fab to bind when the computed rmsd first decreased below 1.5 Å. (In all simulations in which the computed rmsd decreased below 1.5 Å, the Fab–HA binding interface remained stable throughout the remainder of the simulation.) Red x indicates a simulation in which the Fab never bound. (B) Fab rmsd from the crystal structure in the simulation of the CH65–HA complex and one simulation of CH65 binding to HA. The blue curve indicates rmsd fluctuations for CH65 in complex with HA. The red curve shows the rmsd for CH65 as it spontaneously binds to HA in the simulation that corresponds to the one indicated by the orange square in A; rmsd is computed for all backbone atoms in the Fab by first aligning HA in the simulation to HA in the crystal structure (Protein Data Bank ID code 3SM5). (C) Superposition onto the CH65–HA crystal structure of the last frame from one of the simulations of UCA binding to HA. CH65 is shown in light blue, UCA is in yellow, and HA is in the surface representation. D107 of CDR H3 forms polar interactions with HA, which is indicated by the cyan patch on the HA surface. V106 and Y109 of CDR H3 make hydrophobic contacts with HA (highlighted green). Images were created in OpenStructure (37). (D) The Fab rmsd from the crystal structure of the CH65–HA complex in the simulation of the UCA–HA complex (blue) and one simulation of the UCA binding to HA (red); rmsd was computed as in B.
To study whether the UCA binds to HA in the same conformation as the mature antibodies, we carried out a set of 10 simulations, in which we initially displaced a homology model of the UCA based on CH65 by 7 Å from the corresponding bound position of CH65 in the antibody–HA complex. In three of these simulations, the UCA spontaneously bound to HA with a backbone rmsd of less than 1.5 Å from the crystallographic position of CH65; the times to achieve binding were 0.004, 0.073, and 0.175 μs. The contacts between the CDR H3 and the receptor-binding pocket of HA are the same for the UCA and CH65 (shown in Fig. 3C), which superimposes the last conformation of one such binding simulation (Movie S2) on the crystal structure of CH65–HA complex. Fig. 3D shows the time trace of this binding simulation. We also carried out a simulation of the homology model of the UCA in complex with HA, in which the complex remained stable over the full simulation period of 25 μs (Fig. 3D). Thus, our simulations suggest that the UCA binds to HA in essentially the same conformation as the mature antibodies.
We compared MD simulations of four antibodies from the same lineage—UCA, I-2, CH65, and CH67—in complex with HA and as free antibodies. Simulations of the complexes suggest that all four antibodies bind to HA in the same conformation. To characterize the conformations of CDR H3, we grouped the CDR H3 conformations in all our simulations into 10 clusters—including both the complexes and the free antibodies (Figs. S3 and S4). Fig. S3 shows the time series of cluster assignments for each simulated complex. The CDR H3 loops in all complexes occupy conformations that closely resemble one another and exhibit the same contacts with HA (Fig. S4). We refer to CDR H3 conformations with high probability of appearing in the complex of a given antibody as the bound conformations for that antibody.
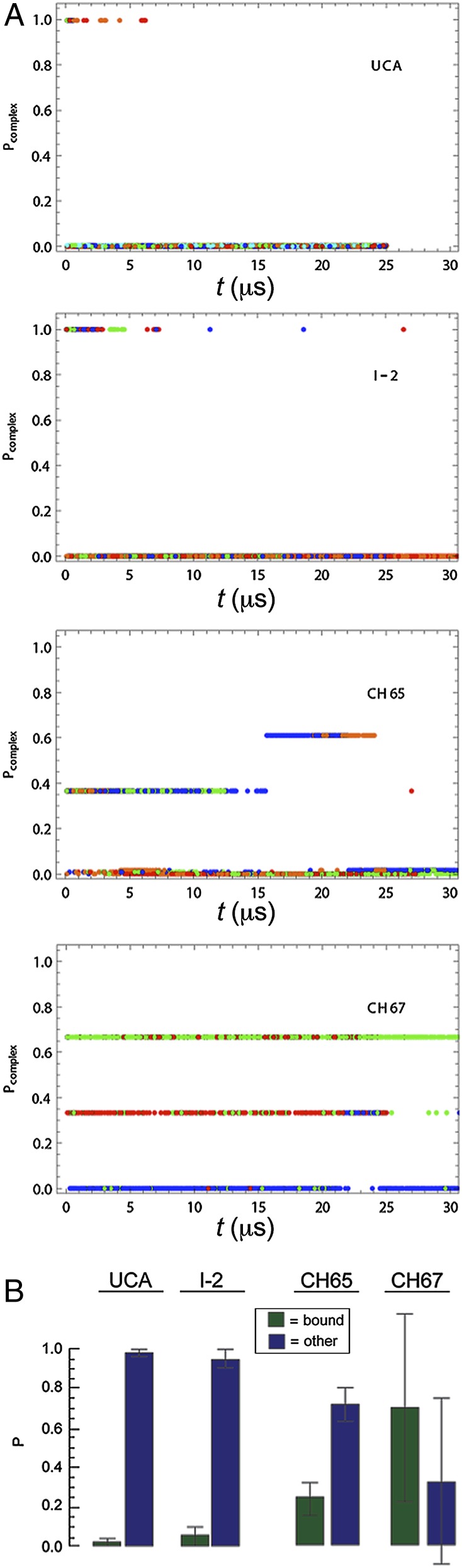
Our simulations of the free antibodies suggest that preconfiguration of the CDR H3 loop accompanies affinity maturation. To illustrate the connection between affinity maturation and the change in CDR H3 conformational dynamics, we associate each CDR H3 conformation observed in these simulations with the probability, Pcomplex, that the conformation is observed in the corresponding complex simulation, and we plot the time trace of these probabilities in Fig. 4A. In all simulations of the UCA, the Fab only briefly occupies conformations with high values of Pcomplex and quickly settles into a variety of different conformations with very low probabilities of appearing in the complex (Movie S3). In contrast, the mature antibodies consistently visit conformations that have high probabilities of appearing in the corresponding complexes.
Fig. 4.
Conformational dynamics of CDR H3 in the simulations of free antibodies. (A) Time trace of the probability that the conformation is observed in the corresponding simulation of the complex. High values indicate that the CDR H3 loop is conformation ready for binding to HA; low values indicate otherwise. Different colors represent different independent simulations. (B) Probability, in the free Fab simulations, that the CDR H3 loop assumes bound and other conformations.
Fig. 4B shows the probability of CDR H3 in the free Fab assuming the corresponding bound conformations and the other conformations. For the UCA and I-2, CDR H3 in the free Fab has a very low probability of assuming the bound conformations; in contrast, for CH65 and CH67, CDR H3 in the free antibody has substantially higher occupancy of the bound conformations. These results suggest that affinity maturation has preconfigured the CDR H3 loop in its bound conformations, decreasing the conformational free-energy penalty in binding and thus, increasing affinity of the antibody for HA.
The initial simulations leading to these conclusions were carried out before the binding experiments and the structures of the free Fabs had been determined. Thus, the MD results anticipated the observation that a slower association rate accounts for weaker binding of the UCA and the conclusion that a principle consequence of the mutations selected during affinity maturation is preconfiguration of the CDR H3 loop.
Discussion
Previous analyses of likely affinity maturation pathways have relied on either related murine monoclonal antibodies against hen egg white lysozyme or other model antigen (9) on comparison of germ-line with mature forms of murine catalytic antibodies (10) or on computational simulations. Several of the earlier studies suggested conclusions similar to our conclusions (11), and computational design efforts led to a proposal that conformational flexibility is an intrinsic property of germ-line–specified CDR H3 sequences (12). Only with the B-cell sorting, variable-region cloning, and antibody expression methods implemented here, however, has it become possible to study a well-defined example of affinity maturation in a human subject responding to a vaccine and analyze multiple stages and branches of the maturation pathway.
The nearly equivalent neutralization profiles of CH65 and CH67 show that these two mature antibodies have acquired similar properties by divergent routes. I-2, the last intermediate shared by CH65 and CH67, differs from the UCA at only three positions; the nucleotide sequences show that a fourth amino acid change, R29, arose independently in the two branches of the lineage. In both branches, increased conformational restriction of CDR H3 has been the principle consequence of affinity maturation. Mutations at different positions in CH65 and CH67 must account for the similar outcomes and in neither case have those changes been in the CDR H3 itself. Early work on kinetics of antibody binding led to a suggestion that flexibility of germ-line–specified complementarity-determining loops might increase the adaptability of the B-cell immune response by allowing the course of affinity maturation to refine the specificity of antigen binding in addition to its strength (13). The CH65–CH67 lineage shows that, if they start from a suitable progenitor, quite different pathways can lead to essentially identical specificities while retaining considerable breadth of recognition of related antigens.
Proposals for novel vaccine strategies that might yield greater breadth of immunity than current seasonal immunizations (universal flu vaccines) include the notion that an immunogen with higher affinity for the ancestor of a known broadly neutralizing antibody would have a greater likelihood of eliciting the desired broad response (14–18). If preconfiguration of the antigen combining site is the principal consequence of affinity maturation, which was shown here for the CH65–CH67 lineage, then making such an immunogen would require compensating for antibody combining site flexibility by introducing into the immunogen interactions that slow dissociation. Determining whether any of the somatic mutations introduced during affinity maturation had a selective effect on the dissociation rate of the Fab–antigen complex could indicate where to create such compensatory interactions. Structural information from mature antibody complexes could facilitate design or selection of such changes in the UCA-contacting surface of an HA-derived immunogen if, as our simulations show for this lineage, the progenitor antibody binds antigen in essentially the same conformation as the mature antibodies.
CH65 and CH67 illustrate that a naive response can evolve in more than one way to increase affinity for antigen (and hence, fitness under conditions of exposure to antigen in the competitive environment of a germinal center). A complementary inference—that different ancestral BCRs can converge to similar binding and neutralization properties—comes from studies of broadly neutralizing antibodies that recognize the CD4 binding site of HIV gp120 (19) and studies on influenza antibody repertoires (20). These observations suggest that antibody affinity maturation in human subjects may, indeed, meet some of the criteria necessary for implementing a B-cell lineage immunogen-design approach to a more universal influenza vaccine.
Materials and Methods
Expression and Purification of Fabs.
The heavy- and light-chain variable and constant domains of UCA, CH65, and CH67 Fabs were amplified from the pcDNA 3.1 expression vector and cloned into the pVRC8400 expression vector using NotI and NheI restriction sites and the tissue plasminogen activator signal sequence. The C terminus of the heavy-chain constructs contained a noncleavable 6xHis tag. I-2 was created using the UCA heavy-chain DNA as a template and mutated at positions G31D, M34I, and N52H by QuikChange Mutagenesis (Agilent) following the manufacturer’s suggested protocol. Fabs were produced by transient transfection of 293T cells using Lipofectamine 2000 (Invitrogen) following the manufacturer’s suggested protocol. Supernatants were harvested 5 d later and clarified from cellular debris by low-speed centrifugation. Fabs were purified using Ni-NTA agarose (Qiagen) followed by gel filtration chromatography on a Superdex 200 column (GE Healthcare).
Expression and Purification of HA Head.
Codon-optimized cDNA of the ectodomain of HA A/Solomon Islands/3/2006 was used as a template to clone the globular head of HA into a pFastBac vector modified for ligation-independent cloning (7). This construct contained an R226Q mutant from the previously reported structure (Protein Data Bank ID code 3SM5). The construct contained a secretion signal at its N terminus and a PreScission cleavage site preceding a 6xHis tag on its C terminus (Dataset S1). Hi-5 cells were infected with recombinant baculovirus. The supernatant was harvested 72 h later and clarified by centrifugation. The HA head was purified by nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) followed by gel filtration chromatography on a Superdex 200 column (GE Healthcare). The C-terminal 6xHis tag was removed by treatment with PreScission protease (GE Healthcare) and carboxypeptidase and repurified by orthogonal Ni-NTA agarose chromatography.
Crystallization.
The CH67 Fab and the A/Solomon Islands/3/2006 globular head were incubated at a 2:1 molar ratio, and the resulting 1:1 complex was purified from excess Fab by gel filtration chromatography on a Superdex 200 column in 10 mM Tris⋅HCl, 150 mM NaCl, pH 7.5. The complex was concentrated to ∼8 mg/mL. Crystals, grown in hanging drops over a reservoir of Tris (pH 8.0) and 46% PEG 400 at 24 °C, appeared within 7 d. They were harvested directly from the drop and flash-cooled in liquid nitrogen. Unbound CH67 Fab was purified as described above and crystallized in hanging drops over a reservoir of 10 mM Tris⋅HCl and 15–20% PEG 4000 at 24 °C. Crystals, which appeared within 3–5 d, were harvested and cryoprotected by the addition of 10–15% PEG 400 or glycerol to the reservoir solution; then, they were flash-cooled in liquid nitrogen. Unbound UCA was purified as described above and crystallized in hanging drops over a reservoir solution of 1.6 M ammonium sulfate at 100 mM sodium acetate at pH 4.8–5.0 at 24 °C. Crystals, which appeared within 3 d, were harvested from the drop and flash-cooled in liquid nitrogen. Unbound I-2 was purified as described above and crystallized in hanging drops over a reservoir solution of 1.5 M sodium citrate at pH 4.9–5.0. Crystals were harvested directly from the drop and flash-cooled in liquid nitrogen. All unbound Fabs were crystallized at ∼10–15 mg/mL.
Structure Determination and Refinement.
We recorded diffraction data at beam lines 24-ID-E at the Advanced Photon Source and 8.2.2 at the Advanced Light Source. Datasets from individual crystals were processed with HKL2000 (21) (Table S1). Molecular replacement (MR) was carried out with PHASER (22), refinement was carried out with PHENIX (23), and all model modifications were carried out with COOT (24). For the CH67–HA complex, we separated the search model into two fragments: the CH65 heavy- and light-chain variable domains–HA complex and the heavy- and light-chain constant domains (Protein Data Bank ID code 3SM5) (7). Simulated annealing refinement of atomic positions and B factors was followed by Translation Libration Screw-motion (TLS) and positional refinement and placement of water molecules. CDR L2 was disordered and not included in the final model. For the UCA Fab, we used the variable and constant domains from the CH65–HA complex (Protein Data Bank ID code 3SM5) as search fragments for MR, removing the CDR H3 loop from the initial model. There are two copies of the UCA Fab in the asymmetric unit (ASU). The CDR H3 loop is disordered in one copy in the ASU; it is at a crystal contact in the other copy, and we could rebuild it in a conformation quite different from its conformation in the CH65–HA and CH67–HA complexes. Initial rigid body refinement was followed by simulated annealing refinement of positions and B factors, with a final TLS and positional refinement and water placement. We applied noncrystallographic symmetry restraints throughout. For the CH67 Fab, we used an initial search model with CH67 Fab from the previously determined higher-resolution CH67–HA complex, removing the CDR H3 loop. Strong electron density for this loop, in the conformation seen in the complex, was present at each of the six copies in the ASU, and we, therefore, included it in a second round of MR. We then rebuilt the model with PHENIX AutoBuild (25) and refined it by simulated annealing with cycles of positional and group B-factor refinement followed by TLS and positional refinement, imposing noncrystallographic symmetry, secondary-structure, and reference-model restraints (using the CH67 Fab from the HA complex) at all stages. As in the CH67–HA complex, the CDR L2 loop was disordered in each copy of the CH67 Fab. Portions of the heavy- and light-chain constant modules had high B factors and lacked clear electron density, and we removed these strands from the model. For the I-2 structure, we used the UCA variable and constant modules as search models, rebuilt the fit with PHENIX AutoBuild, and refined the model by simulated annealing and cycles of positional and B-factor refinement followed by TLS. There was strong density for the heavy- and light-chain variable domains, but the constant module appeared to be flexible, and some regions lacked clear density. The density did not improve during refinement; disorder in the constant module has been described for a number of other Fabs (26). All structures were validated with the MolProbity server (27). Coordinates and diffraction data have been submitted to the Protein Data Bank (ID codes 4HK0, 4HK3, 4HKB, and 4HKX).
Surface Plasmon Resonance Measurements.
Analyses were performed using either a Biacore 3000 or a T100. The HA head was coupled to a CM5 sensor chip, and purified Fab was injected at a flow rate of 30 μL/min. The Biacore 3000 running buffer was 10 mM Hepes, 150 mM NaCl, and 0.005% P20 (pH 7.4). The T100 buffer had a P20 concentration of 0.05%, and it was supplemented with 3 mM EDTA. All sensorgrams were corrected for nonspecific binding with readings from a protein-free reference flow cell. The signal from a buffer-only injection in the active flow cell was subtracted before data evaluation. Data were processed initially using either BiaEval software version 2.0 or the T100 Evaluation software (Biacore), but subsequent fitting was carried out as described below. Regeneration of the CM5 chip was by a brief (10–20 s) pulse of 2 M MgCl2. The binding kinetics of mature antibodies could be fit relatively well with a 1:1 Langmuir model, but the binding kinetics of the UCA and I-2 showed evidence of multiple components. These data were fit with a model as described in SI Materials and Methods and Fig. S5.
Infectivity Neutralization.
A microneutralization assay was performed based on the methods of the influenza reference laboratories at the Centers for Disease Control and Prevention as previously described (7). The effective concentrations of full-length antibody needed to inhibit at least 99% of viral infectivity are reported in Table 1.
DNA Sequencing.
RNA extraction from 140 µL virus was performed using the Qiamp Viral RNA Mini kit (Qiagen) according to the manufacturer’s suggested protocol; 2 µL extracted RNA served as template to make cDNA using the Monsterscript First-Strand cDNA Synthesis Kit (Epicentre Biotechnologies) with the primer Uni12 (5′-AGCAAAAGCAGG-3′), and 2 µL cDNA were used to amplify the HA gene using the Platinum PCR Supermix High Fidelity enzyme (Invitrogen). For each HA gene, two reactions were set up. One reaction used the forward primer BsmBI-HA-1 (5′-TATTCGTCTCAGGGAGCAAAAGCAGGGG-3′, where the underlined nucleotides encode the BsmBI restriction site) and reverse primer 743 (5′- CAATGAAACCGGCAATGGCTCCAAAC-3′). The second reaction used the forward primer 742 (5′-GTTTGGAGCCATTGCCGGTTTCATTG-3′) and the reverse primer BsmBI-HA-R (5′- ATATCGTCTCGTATTAGTAGAAACAAGGGTGTTTT-3′, where the underlined nucleotides encode the BsmBI restriction site). Primers 742 and 743 were designed based on the conserved HA2 fusion peptide region in H1N1 strains. PCR products were purified using the QIAquick PCR purification kit (Qiagen) and submitted to Genewiz for Sanger Sequencing using the same primers used for amplification. Sequence data were analyzed using the Sequencher 4.0 software. The sequences reported in this paper were deposited in GenBank (accession nos. JX477156–JX1477164; nine total).
MD Simulations.
All simulations used the AMBER99SB-ILDN force field (28) for proteins and ions and the TIP3P model for water (29). The AMBER99SB-ILDN force field has improved the side-chain torsion potentials in the AMBER99SB force field (30), which has improved the backbone parameters in the AMBER force field (31). All bond lengths to hydrogen atoms were constrained using M-SHAKE (32). van der Waals and short-range electrostatic interactions were cut off at 10 Å, and long-range electrostatic interactions were calculated using the Gaussian split Ewald method (33). The simulation time step was 2.5 fs; long-range electrostatics were evaluated every third step. A version of the MTK algorithm (34) was used to maintain the simulations in the isobaric–isothermal ensemble with a temperature of 310 K and a pressure of 1 atm. The Higher Order Harmonics integrator (35) was used to propagate the equations of motion. Simulations were performed on a 256- or 512-node special-purpose machine, Anton (8), designed for MD simulations.
The antibodies and the antibody–HA complexes were first equilibrated for 50 ns with their backbone atoms restrained by a harmonic potential, which was gradually turned off during the course of the equilibration. Replicate simulations of free antibodies were initiated in conformations extracted at 5-μs intervals from the corresponding antibody–HA simulations. Homology models were constructed by mutating the involved residues in Maestro (36).
Supplementary Material
Acknowledgments
We thank James Whittle, Junhua Pan, and Marco Morelli for help with cloning and purification of HA head construct and the beam line staff at Advanced Photon Source 24-ID-E and Advanced Light Source 8.2.2. We also thank Ansgar Philippsen for creating some of the molecular graphics and figures. Research reported in this publication was supported by National Institute of Allergy and Infectious Diseases of the National Institutes of Health Grant P01AI089618.
Footnotes
The authors declare no conflict of interest.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4HK0, 4HK3, 4HKB, and 4HKX). The sequences reported in this paper have been deposited in the GenBank database (accession nos. JX477156, JX477157, JX477158, JX477159, JX477160, JX477161, JX477162, JX477163, and JX477164).
See Commentary on page 7.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218256109/-/DCSupplemental.
References
- 1.Eisen HN, Siskind GW. Variations in affinities of antibodies during the immune response. Biochemistry. 1964;3:996–1008. doi: 10.1021/bi00895a027. [DOI] [PubMed] [Google Scholar]
- 2.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 3.McKean D, et al. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc Natl Acad Sci USA. 1984;81(10):3180–3184. doi: 10.1073/pnas.81.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liao HX, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods. 2009;158(1–2):171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wardemann H, et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301(5638):1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 6.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453(7195):667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whittle JR, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci USA. 2011;108(34):14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw DE, et al. 2009. Millisecond-scalemolecular dynamics simulations on Anton. Proceedings of the Conference on High Performance Computing, Networking, Storage and Analysis (SC09) (Association for Computing Machinery, New York)
- 9.Braden BC, Goldman ER, Mariuzza RA, Poljak RJ. Anatomy of an antibody molecule: Structure, kinetics, thermodynamics and mutational studies of the antilysozyme antibody D1.3. Immunol Rev. 1998;163:45–57. doi: 10.1111/j.1600-065x.1998.tb01187.x. [DOI] [PubMed] [Google Scholar]
- 10.Wedemayer GJ, Patten PA, Wang LH, Schultz PG, Stevens RC. Structural insights into the evolution of an antibody combining site. Science. 1997;276(5319):1665–1669. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- 11.Patten PA, et al. The immunological evolution of catalysis. Science. 1996;271(5252):1086–1091. doi: 10.1126/science.271.5252.1086. [DOI] [PubMed] [Google Scholar]
- 12.Babor M, Kortemme T. Multi-constraint computational design suggests that native sequences of germline antibody H3 loops are nearly optimal for conformational flexibility. Proteins. 2009;75(4):846–858. doi: 10.1002/prot.22293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foote J, Milstein C. Kinetic maturation of an immune response. Nature. 1991;352(6335):530–532. doi: 10.1038/352530a0. [DOI] [PubMed] [Google Scholar]
- 14.Dal Porto JM, Haberman AM, Kelsoe G, Shlomchik MJ. Very low affinity B cells form germinal centers, become memory B cells, and participate in secondary immune responses when higher affinity competition is reduced. J Exp Med. 2002;195(9):1215–1221. doi: 10.1084/jem.20011550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dal Porto JM, Haberman AM, Shlomchik MJ, Kelsoe G. Antigen drives very low affinity B cells to become plasmacytes and enter germinal centers. J Immunol. 1998;161(10):5373–5381. [PubMed] [Google Scholar]
- 16.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol. 2012;30(5):423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwickert TA, et al. A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. J Exp Med. 2011;208(6):1243–1252. doi: 10.1084/jem.20102477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shih TA, Meffre E, Roederer M, Nussenzweig MC. Role of BCR affinity in T cell dependent antibody responses in vivo. Nat Immunol. 2002;3(6):570–575. doi: 10.1038/ni803. [DOI] [PubMed] [Google Scholar]
- 19.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333(6049):1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krause JC, et al. Epitope-specific human influenza antibody repertoires diversify by B cell intraclonal sequence divergence and interclonal convergence. J Immunol. 2011;187(7):3704–3711. doi: 10.4049/jimmunol.1101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-Ray Diffraction Data Collected in Oscillation Mode. London: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 22.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 1):61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324(5924):246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorgensen WL, et al. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 30.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cornell WD, et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 32.Kräutler V, van Gunsteren WF, Hünenberger PH. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J Comput Chem. 2001;22:501–508. [Google Scholar]
- 33.Shan Y, Klepeis JL, Eastwood MP, Dror RO, Shaw DE. Gaussian split Ewald: A fast Ewald mesh method for molecular simulation. J Chem Phys. 2005;122(5):54101. doi: 10.1063/1.1839571. [DOI] [PubMed] [Google Scholar]
- 34.Martyna GJ, et al. Explicit reversible integrators for extended systems dynamics. Mol Phys. 1996;87:1117–1157. [Google Scholar]
- 35.Predescu C. Computationally efficient molecular dynamics integrators with improved sampling accuracy. Mol Phys. 2012;110:967–983. [Google Scholar]
- 36.Schrodinger . (2011) Maestro, (Schrodinger, LLC, New York), Version 9.2. [Google Scholar]
- 37.Biasini M, et al. OpenStructure: A flexible software framework for computational structural biology. Bioinformatics. 2010;26(20):2626–2628. doi: 10.1093/bioinformatics/btq481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.