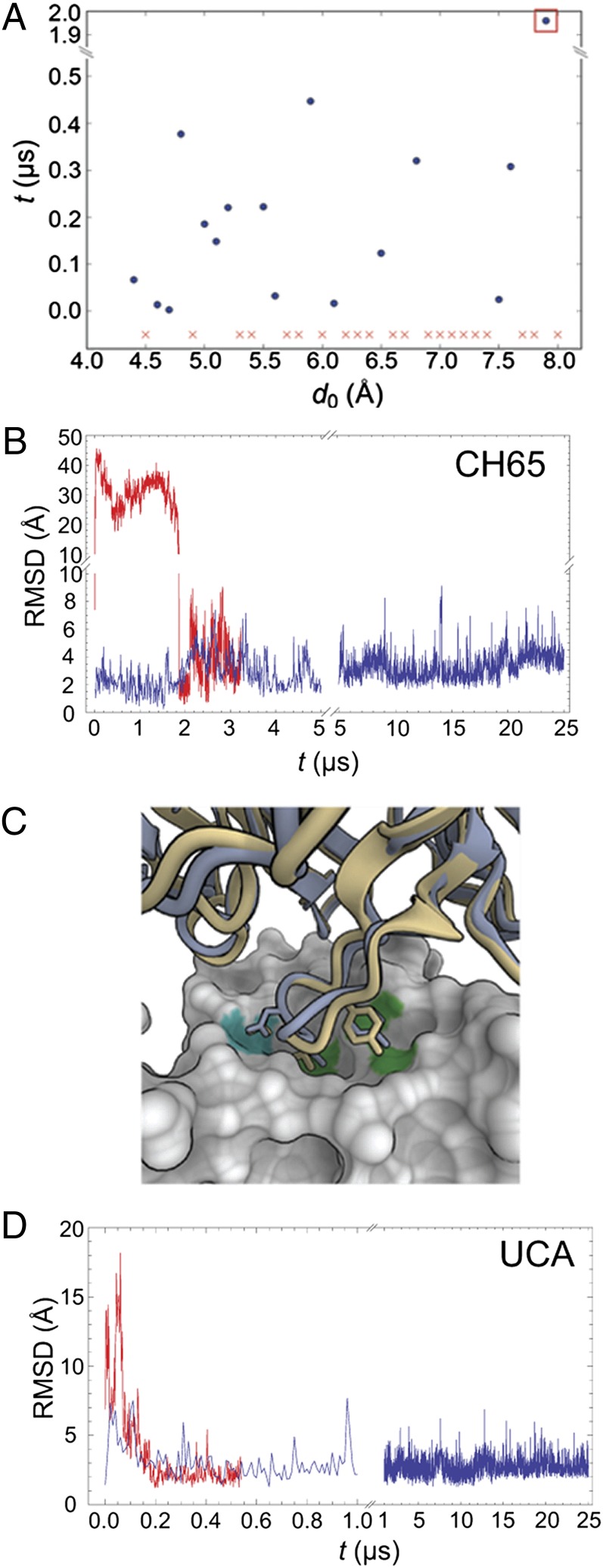
Fig. 3.
Long time-scale MD simulations. (A) Time required for the CH65 Fab to bind spontaneously to HA starting from various initial separations. Each of these simulations was initiated with the CH65 Fab placed at a different distance, d0, from its position within the CH65–HA complex. Details of these simulations are described in SI Materials and Methods. Each simulation was 0.5 μs in length, except for those simulations with d0 > 7.5 Å, which were 5 μs long. To determine whether the Fab had bound to HA, we aligned HA to the CH65–HA crystal structure, computed the Fab backbone rmsd with respect to its position in the complex, and considered the Fab to bind when the computed rmsd first decreased below 1.5 Å. (In all simulations in which the computed rmsd decreased below 1.5 Å, the Fab–HA binding interface remained stable throughout the remainder of the simulation.) Red x indicates a simulation in which the Fab never bound. (B) Fab rmsd from the crystal structure in the simulation of the CH65–HA complex and one simulation of CH65 binding to HA. The blue curve indicates rmsd fluctuations for CH65 in complex with HA. The red curve shows the rmsd for CH65 as it spontaneously binds to HA in the simulation that corresponds to the one indicated by the orange square in A; rmsd is computed for all backbone atoms in the Fab by first aligning HA in the simulation to HA in the crystal structure (Protein Data Bank ID code 3SM5). (C) Superposition onto the CH65–HA crystal structure of the last frame from one of the simulations of UCA binding to HA. CH65 is shown in light blue, UCA is in yellow, and HA is in the surface representation. D107 of CDR H3 forms polar interactions with HA, which is indicated by the cyan patch on the HA surface. V106 and Y109 of CDR H3 make hydrophobic contacts with HA (highlighted green). Images were created in OpenStructure (37). (D) The Fab rmsd from the crystal structure of the CH65–HA complex in the simulation of the UCA–HA complex (blue) and one simulation of the UCA binding to HA (red); rmsd was computed as in B.