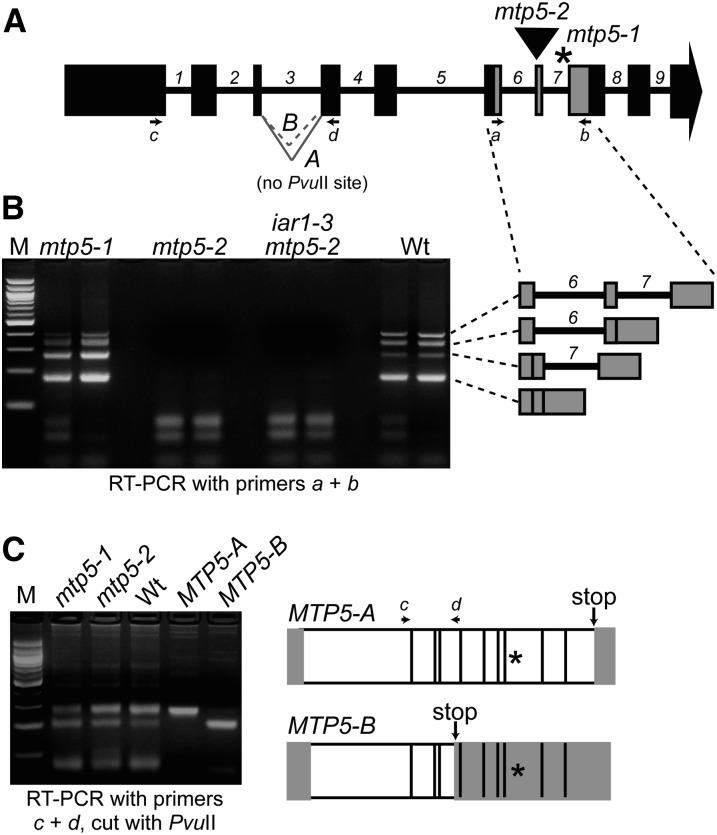
Figure 4 .
Alternative splicing of MTP5 transcripts. (A) Model of MTP5 gene showing exons (boxes), introns (numbered), positions of mtp5 mutations (asterisk, mtp5-1; triangle, mtp5-2), primers used for RT-PCR amplification (arrows a-d), and the A and B MTP5 splice variants of intron 3. (B) mtp5-2 seedlings lack intact MTP5 transcript, and mtp5-1 and wild-type seedlings inefficiently splice MTP5 introns 6 and 7. MTP5 transcripts were detected via RT-PCR amplification using gene-specific primers (A and B) flanking both mtp5 mutations with RNA isolated from 7-d-old mtp5-1, mtp5-2, iar1-3 mtp5-2, and Col-0 (Wt) seedlings. Adjacent lanes show PCR-amplification products of cDNA from two independent reverse transcription reactions using the same RNA. The identities of the four amplicons from the RT-PCR analysis using primers a and b were determined by sequencing and are shown to the right in gray. (C) mtp5-1 and wild-type seedlings alternatively splice MTP5 intron 3. Using gene-specific primers c and d (A), MTP5 transcripts were amplified from cDNA reverse-transcribed from Col-0 (Wt), mtp5-1, and mtp5-2 RNA. RT-PCR analysis revealed two transcripts in wild type and the two mutant alleles, designated MTP5-A and MTP5-B. Digesting the PCR products with PvuII distinguished MTP5-A, which lacks a PvuII site, from MTP5-B, which includes a PvuII site (agarose gel image in C). Cloned MTP5-A and MTP5-B cDNAs were included as positive controls. The transcripts were differently spliced at the 3′ end of the third intron, resulting in a premature stop codon (arrows) in MTP5-B. Asterisks mark the position of the mtp5-1 mutation. White boxes indicate the open reading frame of the resultant transcripts, and gray boxes show predicted untranslated regions.