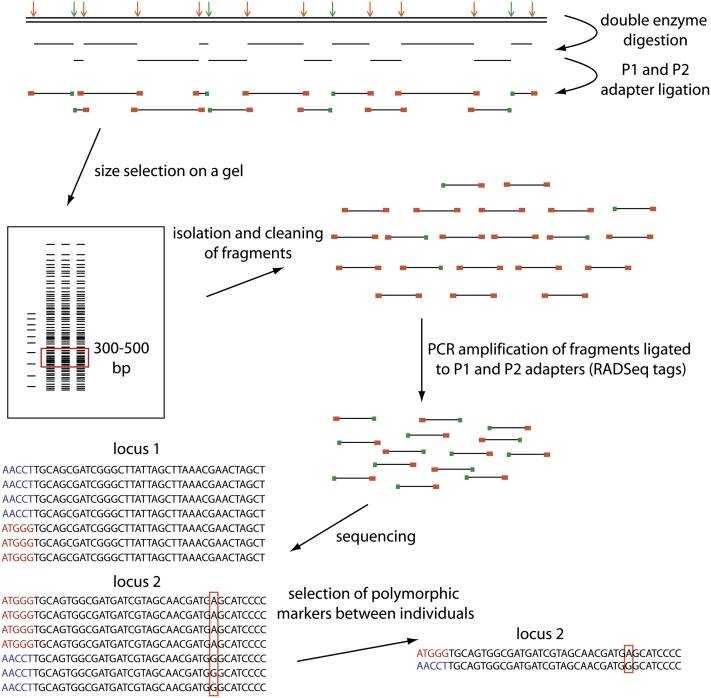
Figure 1 .
Schematic of the ddRADSeq protocol. Arrows indicate restriction sites for a rare-cutting enzyme in green and a frequent-cutting enzyme in orange. P1 adapters specifically bind to the rare-cutting enzyme (green) and P2 adapters bind to the frequent-cutter (orange) restriction sites. Each individual contains a different barcode of five nucleotide bases specified by the P1 adapter. Sequenced RAD tags start with the barcode (individual one: red barcode, individual two: blue barcode). The bioinformatic software Stacks allows for the detection of single nucleotide polymorphisms between individuals (outlined by red box in sequence alignment).