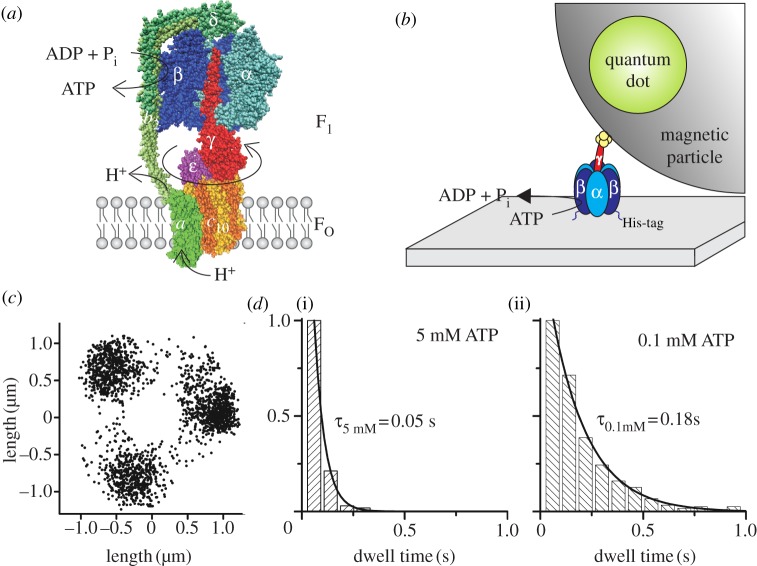
Figure 1.
(a) Structural model of E. coli FOF1-ATP synthase with one α (light blue) and one β (dark blue) subunit removed to expose subunit γ (red) in the centre of the α3β3-pseudohexagon. Together with ɛ (pink) and δ on top (dark green), they form the F1 portion. The membrane-embedded FO portion consists of 10 c subunits (yellow/orange), which form a ring structure within the membrane. Subunit a (light green) and two b subunits (green) form an eccentric stalk to hold both portions together. Subunits γ, ε and the c10-ring comprise the rotor of FOF1 (reddish colours), which rotates clockwise (if viewed from the membrane) during ATP synthesis with respect to the stator subunits α3β3δab2 (blue/green colours). ATP synthesis in β is driven by proton (or Na+) flow through FO owing to an electrochemical potential difference over the membrane. This homology model of FOF1 is a composite of several partial structures (PDB codes 1BMF [16], 2CK3 [30], 1H8E [31] and 1YCE [32] in the Protein Data Bank, http://www.pdb.org) and was drawn with VMD v. 1.9.1 [33]. Subunits a, b2 and δ have been modelled in roughly their correct size and manually attached using cross-link data. (b) Set-up of the rotation assay with a magnetic bead attached to the F1 portion. F1 (α3β3γ) is attached to a cover glass via histidine (His)-tags in each β subunit. The streptavidin-covered magnetic bead is coupled to a cysteine in subunit γ via biotin. The bead is doped with fluorescent biotinylated quantum dots to visualize ATP-driven rotation of the γ-bead complex. (c) Endpoint distribution of a rotating actin filament (1 µm) coupled to the c10-ring of E. coli FOF1 in the rotation assay showing apparent 120° stepping rotation during ATP hydrolysis at high [ATP] in the presence of detergent (adapted from [34]). (d) Dwell times of stepped rotation of E. coli FOF1 in the presence of (i) 5 mM or (ii) 0.1 mM ATP. The histograms show the averaged dwells of the stepping positions of one enzyme after 200 or 120 rotations, respectively.