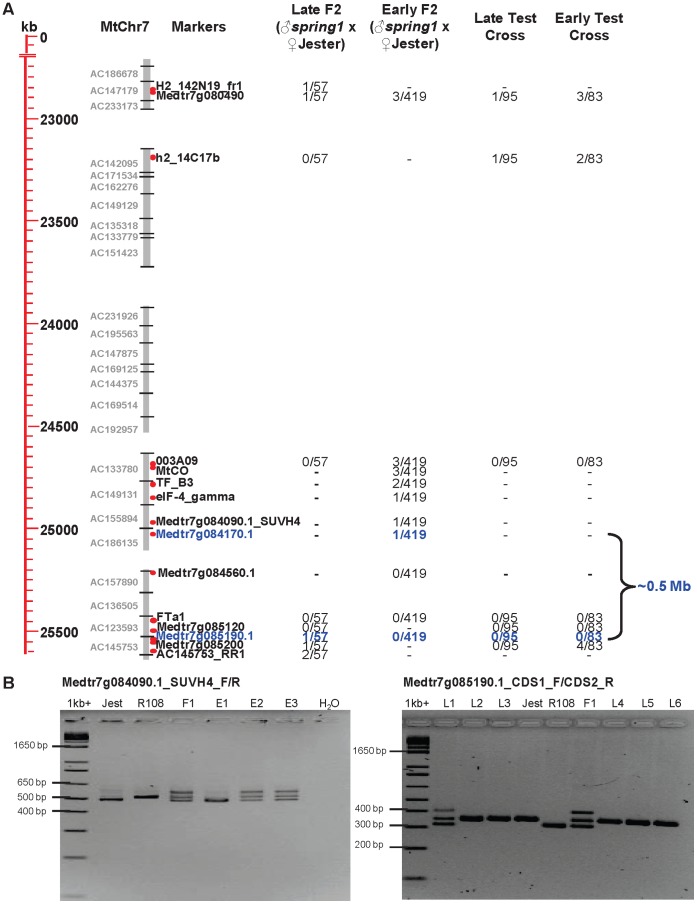
Figure 4. Markers for fine mapping spring1 on chromosome 7 and defining the ∼0.5 Mb interval that contains spring1.
a) Physical map of the spring1 region on chromosome 7 with DNA sequence in kilobases (kb), Bacterial Artificial Chromosome (BAC) clone contigs (grey bars) and mapping marker position (red dots). Markers defining the spring1 interval are blue. Four columns show the numbers of recombinants detected with the markers in the early and late flowering plants from the two Jester mapping populations; the F2 plants from cross “spring1 x Jester” and the progeny of the Test cross. b) Examples of PCR genotyping using two indel DNA markers flanking the spring1 interval. Control PCR reactions from Jester (Jest), R108 and F1 from the control cross (“R108 x Jester”) are shown. R108 and spring1 gave the same PCR products in all cases. PCR genotyping with marker Medtr7g084090.1 (left). Products from genotyping of three early flowering F2 plants (E1 to E3) from the Mapping cross “spring1xJester”. Plant E1 is homozygous for the Jester band, thus Medtr7g084090.1 is separated from spring1 by a recombination event. Genotyping with marker Medtr7g085190.1 on six late flowering F2 plants (L1 to L6) from the Mapping cross “spring1xJester” (right). Plant L1 is heterozygous, thus Medtr7g085190.1 is separated from spring1 by a recombination event. A feature of both indel markers is the F1 plants and the heterozygous plants give three bands after PCR. These are the expected Jester and R108 bands and a third larger band which is likely to be a heteroduplex of the two PCR differently-sized fragments that is slightly retarded during gel electrophoresis compared to the other bands. PCR products were separated by electrophoresis on a 3% agarose gel and photographed. The Invitrogen 1 kb+ ladder provided molecular size standards. Physical maps were redrawn from a Chromosome Visualisation Tool (CViT) BLAST search http://medicagohapmap.org/with the marker sequences against the current Medicago pseudomolecule Mt3.5 genome assembly http://blast.jcvi.org/er-blast/index.cgi?project=mtbe.