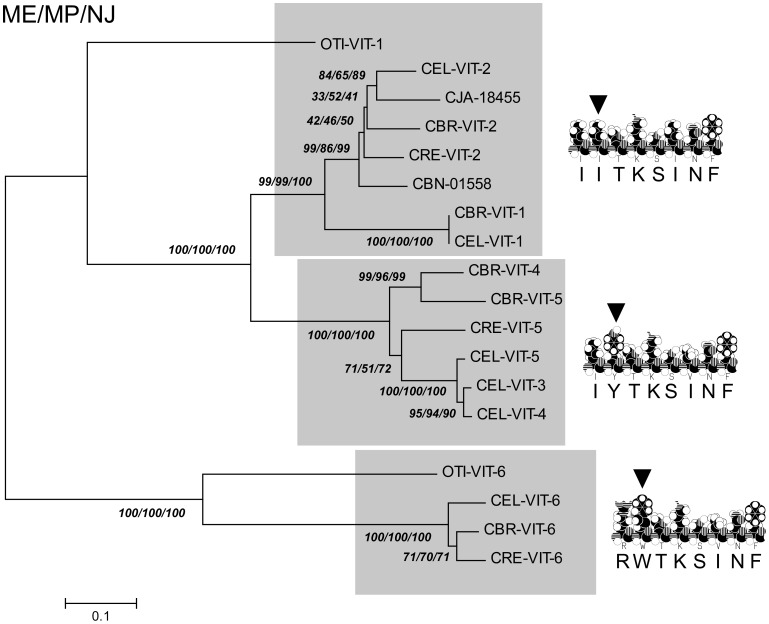
Figure 7. Phylogenetic analysis of the LLT module of a nematode vitellogenin amino acid sequence.
The LLT module of all fifteen yolk protein sequences known from Caenorhabditis spp. and Oscheius tipulae were aligned with Clustal X, using the Blosum64 score matrix and the default values of the software for gap penalties. The alignment was subjected to phylogenetic analysis using Minimum Evolution (ME), Maximum Parsimony (MP) and Neighbor-Joining (NJ) methods. The numbers shown in the unrooted tree represent the bootstrap values obtained from 10,000 replicate trees (ME, MP and NJ). The shadowed branches correspond to the amino acid sequences containing the consensus VTGR binding site obtained from Table S2 shown at right of the tree. The bar at the lower left shows the number of substitutions per 100 amino acid residues.