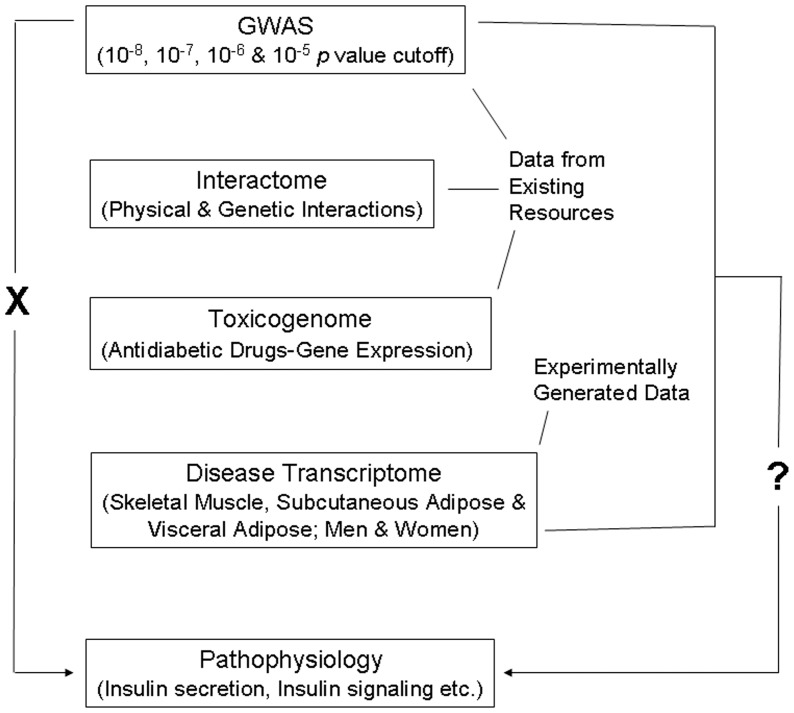
Figure 1. Schematic representation of the workflow.
T2D GWAS genes do not directly relate (indicated by ‘X’ on the left side) to pathways associated with disease pathophysiology. Conspicuously, effect of identified risk variants on continuous glycemic measures in nondiabetic subjects chiefly explains only perturbation of insulin secretion, not insulin resistance. Further, the genes found as associated with the disease do not clearly relate to processes and pathways consistent with the known aspects of T2D pathophysiology. The main aim of the present study was to ask the question (indicated by ‘?’ on the right side) if GWAS data when considered in conjunction with interactome, toxicogenome and disease transcriptome data reveal genome to phenome correlation in T2D. Data available in public domain for GWAS, interactome and toxicogenome was used in the analysis. For disease transcriptome, new experimental data was generated. We specifically examined if interaction network of genes reported in T2D GWAS, genes showing altered expression after treatment with various antidiabetic drugs, and genes that are differentially expressed in insulin responsive tissues in male and female T2D patients do converge on insulin secretion, insulin resistance and other T2D associated pathophysiological pathways.