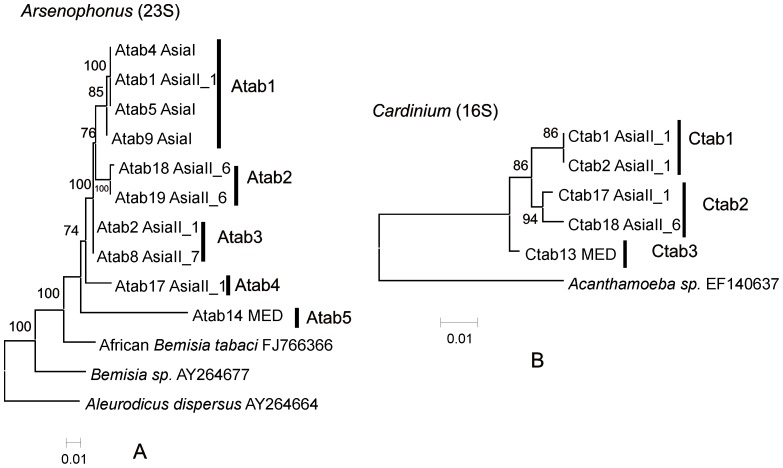
Figure 3. Phylogenies of the S-endosymbionts Arsenophonus and Cardinium.
A. Phylogenetic tree reconstruction based on 23S rRNA gene sequences (length = 550 bp) of Arsenophonus of B. tabaci cryptic species using ML analysis under the HKY+G substitution model. The bootstrap values are indicated. African B. tabaci (FJ66366), Bemisia sp. (AY264677) and Aleurodicus disperses (AY264664) are used as the outgroups. Accession numbers for sequences used in the tree are JX428666–JX428675. All 23S sequences of Arsenophonus used in this study were clustered with other blast references sequences from GenBank and their ML phylogenetic reconstruction is shown Fig. S1E. B. Phylogenetic tree reconstruction based on 16S rRNA gene sequences (length = 400) of Cardinium of B. tabaci cryptic species using MLd analysis under the K2 substitution model. The bootstrap values are indicated. Acanthamoeba sp. (EF140637) is used as the out group. Accession numbers for sequences used in the tree are JX428676–JX428680. All 16S sequences of Cardinium used in this study were clustered with other blast references sequences from GenBank and their ML phylogenetic reconstruction is shown Fig. S1F.