Table 2. Comparison of the missing peak rate of the fixed number-based method (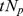 ) and the Benjamini-Hochberg (B–H) algorithm with
) and the Benjamini-Hochberg (B–H) algorithm with 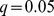 on the 32 spectra of the eight proteins in the benchmark set picked by PICKY.
on the 32 spectra of the eight proteins in the benchmark set picked by PICKY.
| Spectra | 15N-HSQC | HNCO | HNCA | HNCACB | CBCA(CO)NH | ||||||||||
| Protein | tNp | B-H | (d) | tNp | B-H | (d) | tNp | B-H | (d) | tNp | B-H | (d) | tNp | B-H | (d) |
| RP3384 | 6 | 4 | (33) | 0 | 0 | (0) | 13 | 13 | (0) | – | – | 36 | 10 | (72) | |
| CASKIN | 2 | 2 | (0) | 22 | 15 | (32) | – | – | 33 | 38 | (−15) | 10 | 9 | (10) | |
| VRAR | 7 | 7 | (0) | 9 | 9 | (0) | – | – | 31 | 31 | (0) | 19 | 19 | (0) | |
| HACS1 | 5 | 2 | (60) | 6 | 4 | (33) | – | – | 18 | 17 | (6) | 9 | 6 | (33) | |
| TM1112 | 8 | 1 | (88) | – | – | 6 | 6 | (0) | 8 | 7 | (13) | 9 | 2 | (78) | |
| COILIN | 4 | 0 | (100) | 19 | 3 | (84) | – | – | 24 | 24 | (0) | 33 | 20 | (39) | |
| ATC1776 | 5 | 4 | (20) | 8 | 5 | (38) | 19 | 18 | (5) | – | – | 28 | 26 | (7) | |
| YST0336 | 2 | 2 | (0) | 6 | 3 | (50) | 12 | 10 | (17) | – | – | 17 | 12 | (29) | |
| Average | 4.9 | 2.7 | (38) | 10.0 | 5.6 | (34) | 12.5 | 11.7 | (6) | 22.8 | 23.4 | (1) | 20.1 | 13.0 | (34) |
| SDave | 2.0 | 2.0 | 7.2 | 4.6 | 4.6 | 4.4 | 9.1 | 10.8 | 10.3 | 7.5 | |||||
| Preave | 85 | 69 | 77 | 67 | 83 | 70 | 65 | 69 | 68 | 60 | |||||
Column 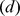 is the relative improvement of the missing peak rate of B-H over
is the relative improvement of the missing peak rate of B-H over 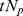 . All values except the last two rows are the missing peak rates. The “
. All values except the last two rows are the missing peak rates. The “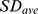 ” row lists the standard deviations of the missing peak rates for the corresponding columns, which demonstrates the robustness of different methods. The last row gives the average precision values. All values are given in percentage.
” row lists the standard deviations of the missing peak rates for the corresponding columns, which demonstrates the robustness of different methods. The last row gives the average precision values. All values are given in percentage.
