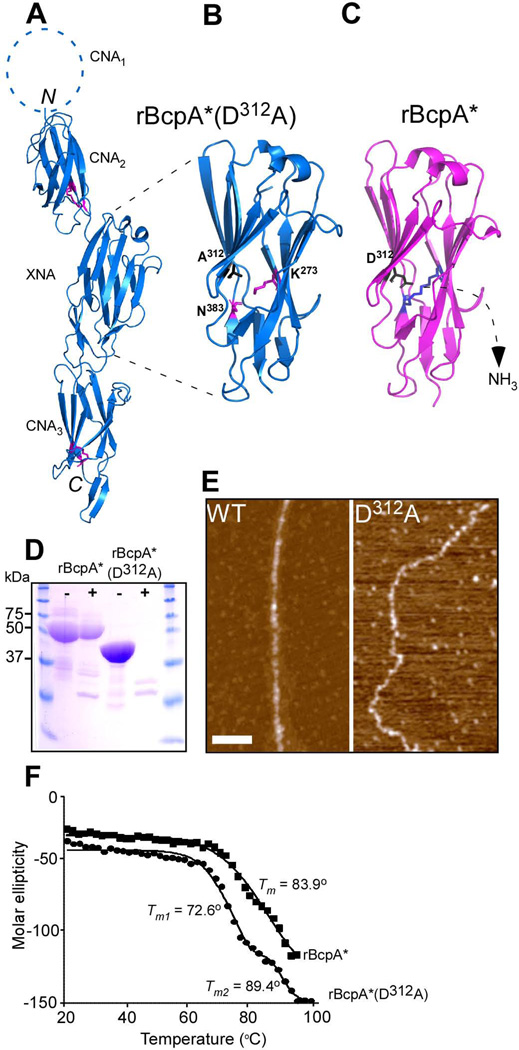
Figure 5.
Crystal structure of rBcpA*(D312A). (A) The crystal structure of rBcpA*(D312A) was solved using molecular replacement. The CNA2, XNA, and CNA3 domains are shown as blue ribbons, the structure of the CNA1 domain is not included in rBcpA*(D312A). The Lys and Asn residues forming the intramolecular isopeptide bonds in the CNA2 and CNA3 domains are shown as black sticks. The intramolecular isopeptide bond in the XNA domain is not formed. The CNA2 and CNA3 domains assume reverse Ig-like folds and the XNA domain a jellyroll fold. (B+C) Close up of the XNA domain of rBcpA*(D312A) (blue) and rBcpA* (magenta), in which the A312 and the catalytic residue D312 are in black and the K273 and N383 residues in magenta and blue and presented as sticks, respectively. In rBcpA*(D312A), the K273-N383 intramolecular isopeptide bond between the two penultimate β-strands is not formed, as the catalyst D312 was replaced with alanine. Formation of the XNA isopeptide bond of rBcpA* (blue sticks) is accompanied by the loss of ammonium. (D) Treatment of rBcpA* and rBcpA*(D312A) with (+) and without (-) trypsin protease revealed that the absence of the XNA isopeptide bond in rBcpA*(D312A) renders the protein sensitive to protease; rBcpA* is protease resistant. (E) AFM images of wild-type (WT) and mutant D312A pili showing the morphological difference between these two types of pili. (F) Thermal melt spectroscopy analysis as a function of temperature demonstrated that rBcpA*(D312A) is less stable than rBcpA*.