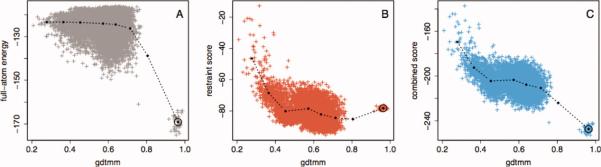
Figure 4.
Full-atom energy and homology-derived spatial restraints distinguish between models in different accuracy regimes. We constructed models for a protein of unknown structure during the CASP9 experiment (CASP9 target T0569). Models were made using the Rosetta rebuild and refine protocol supplemented with the evolutionary restraints as described in Methods section. After obtaining the experimentally determined structure of T0569, we calculated the GDTMM of each model, which approaches 1.0 as a model become more similar to the native (Supporting Information Text 4). The same statistics were calculated for an ensemble of Rosetta refined native structures. Models were assigned to GDTMM bins, which ranged from 0.1 to 1.0 in. increments of 0.1. In each plot, the points connected by lines represent the statistics calculated on each bin, and the gray, red, and blue points represent individual structures. A: Median GDTMM versus median Rosetta full-atom score, with a circle surrounding the bin containing the refined native structures. B: Median GDTMM versus median spatial restraint score. The Rosetta full-atom energy is very effective at discriminating the high-quality from medium-quality models, while less effective at discriminating medium-quality from low-quality models. Conversely, the restraints discriminate medium-quality from low-quality models very well, but are not effective at discriminating high-quality models from natives and can even provide a barrier to sampling the native conformation. C: A combination of the two scores is effective at discrimination independent of model quality.