Figure 4.
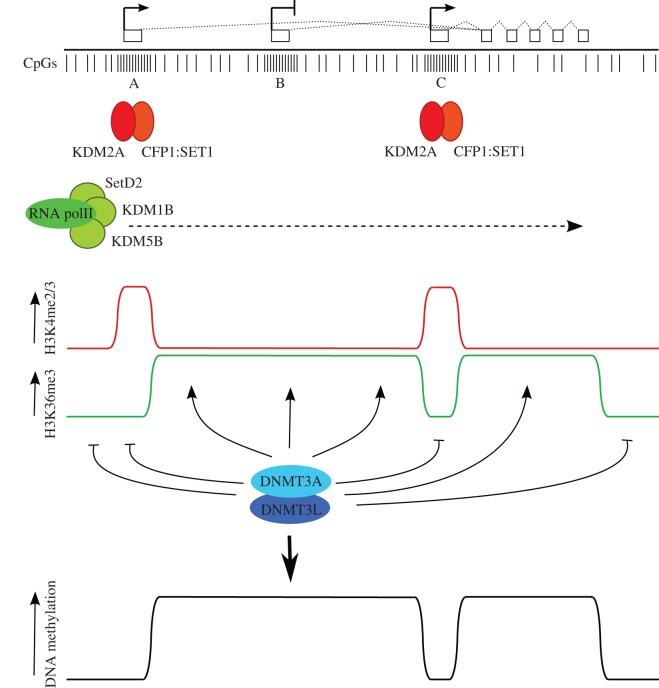
A model for DNA methylation in oocytes incorporating transcription and histone modification state. At the top, an idealized transcription unit active in oocytes, defined by an upstream, oocyte-specific transcription start site (A), and two intragenic CpG island promoter regions, one (B) that is inactive in oocytes, and a second (C) that is expressed in oocytes. Directly below, individual CpG dinucleotides are indicated, with dense clusters of CpGs at CpG islands. CpG islands A and C are predicted to be bound by CxxC-containing proteins KDM2A and CFP1 that have H3K36 demethylase and H3K4 methyltransferase activities, respectively (the latter via recruitment of the SET1 complex). Transcription through the locus brings the H3K36 methyltransferase SETD2 and the H3K4 demethylases KDM5B and KDM1B, as these activities are associated with the elongating RNA polII complex. (Note that these predictions are based on the properties of these proteins established in somatic cells, only KDM1B has thus far been shown to be involved in gDMR methylation in oocytes. Other members of the respective protein families may perform these roles in oocytes.) The combination of these activities generates distinct states of H3K4 and H3K36 methylation within transcribed regions, active and inactive CpG island promoters and intergenic regions. This profile of H3K4 and H3K36 methylation is read by the DNMT3A/DNMT3L de novo DNA methyltransferase complex to establish methylation at gene bodies and silent, intragenic CpG islands and generate the DNA methylation profile indicated at the bottom. By this mechanism, CpG island B could become a gDMR; differential acquisition of DNA methylation of intragenic CpG islands B and C may be determined by promoter activity or other factors that promote binding of proteins such as KDM2A and CFP1 at CpG island C that are hostile to DNA methylation.