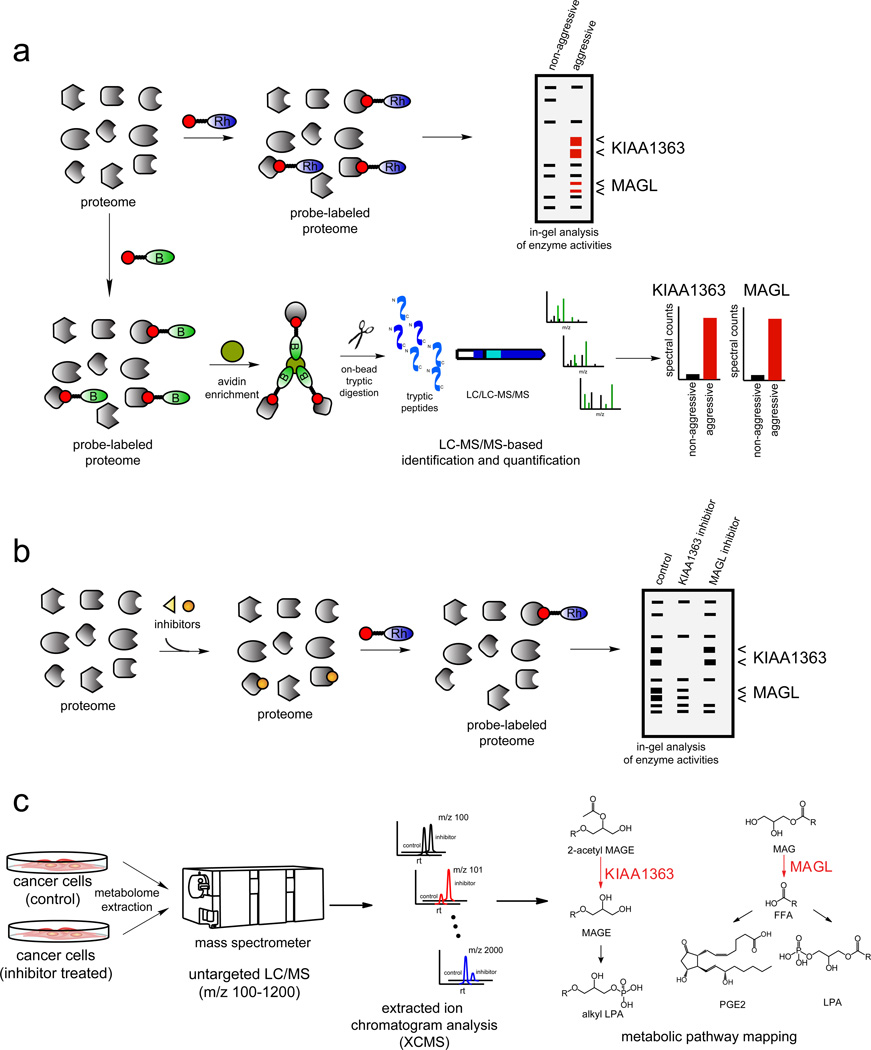
Figure 5. Activity-Based Protein Profiling (ABPP) Coupled with Untargeted Metabolomics in Annotating Dysregulated Enzyme Activities in Aggressive Cancers.
a) ABPP uses active site-directed probes to assess the functional state of large numbers of enzymes directly in complex proteomes (Nomura et al., 2010a). Activity-based probes (ABPs) consist of a reactive group, a spacer arm, and a detection handle (e.g. fluorophore such as a rhodamine (Rh) or biotin (B)). In a typical ABPP experiment, a proteome is reacted with the activity-based probe and readout either by fluorescence on a 1D-SDS-PAGE gel for rhodamine-ABPs (above), or by avidin enrichment, on-bead tryptic digest, and identification and quantification of peptides by Multidimensional Protein Identification Technology (MudPIT) for biotinylated-ABPs (below) (Nomura et al., 2010a). Through ABPP analysis of the serine hydrolase proteome with the serine hydrolase ABP fluorophosphonate (FP)-Rh or FP-biotin, KIAA1363 and MAGL were identified as upregulated in multiple human aggressive cancer cells and primary tumors (Jessani et al., 2002; Nomura et al., 2010b). b) ABPP can also be used in a competitive format to assess potency and selectivity of inhibitors in complex proteomes. Inhibitors can compete with the ABP and enzyme inhibition will be read-out by loss of fluorescence on a SDS-PAGE gel (using a rhodamine-ABP) or loss of spectral counts by mass spectrometry (using a biotintylated-ABP). Competitive ABPP was used to develop selective inhibitors of KIAA1363 and MAGL (Chang et al., 2011b; Chiang et al., 2006; Long et al., 2009). c) With selective inhibitors in hand, the metabolic roles of KIAA1363 and MAGL were elucidated using untargeted LC-MS-based metabolomic approaches in which metabolomes were extracted and analyzed by LCMS, broadly scanning for metabolites across a large mass range. To complement the large amount of data that arises from untargeted metabolomic analysis, powerful software tool XCMS was used for quantitation and identification of ions within LC/MS datasets (Smith et al., 2006) which aligns, quantifies, and statistically rank ions that are altered between two sets of metabolomic data. This methodology was used to annotate KIAA1363 and MAGL as enzymes that regulates ether lipid and fatty acid networks, respectively (Chiang et al., 2006; Nomura et al., 2010b).