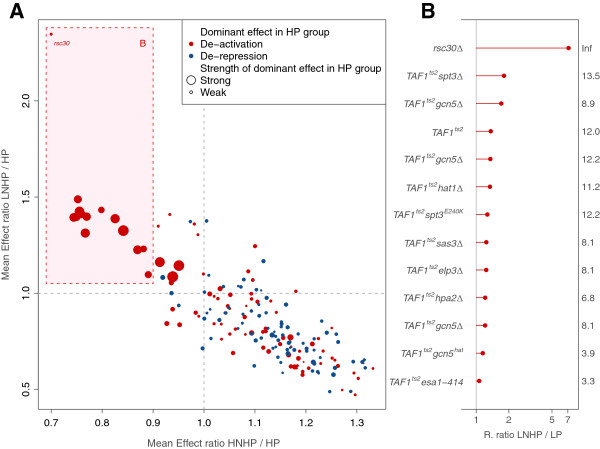
Figure 2.
Distinct chromatin regulation strategies to achieve noisy or quiet plasticity. A) Each dot represents the mean effect in the expression of a set of genes in a subclass (HNHP, x coordinate; LNHP, y coordinate; normalized by effect in HP class) when a particular regulator is mutated [18]. A ratio >1 thus implies that the corresponding subclass is more strongly influenced by certain regulator than the full HP group. A strong negative correlation is found indicating that many regulators are highly specific to either HNHP or LNHP genes. This confirms that these groups are enriched by complementary functional classes (stress and growth related genes, respectively) which are generally regulated in opposite sense [12,13,20]. Dot colors denote the dominant effect of the regulator on the HP class (blue; regulator is mostly repressing expression, red; regulator is commonly activating) while sizes describe the strength of the dominant effect; e.g., LNHP genes are frequently affected by strong chromatin activators. B) We examined in detail the effects on LNHP genes (box in A). Except rsc30 (a regulator of ribosomal proteins [21]) all these mutations involved TAF1, which is part of the general transcription factor TFIID [22,23]. This essential factor regulates ∼90% of the genes in the genome, not including most of HNHP (which are regulated by SAGA) but including almost all LP genes (see main text). Nevertheless, we observed that all these mutations affected significantly more strongly LNHP than LP genes [K-S tests with FDR-corrected -log(p-value)’s shown at the right].