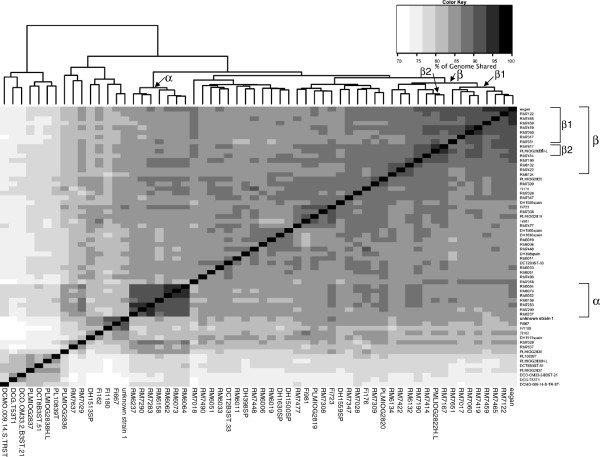
Figure 1.
Whole genome heat map, constructed by Mauve, to achieve pairwise percentage of genome sequence alignment. Pair-wise Mauve alignments were conducted with 60 H. influenzae and H. haemolyticus genome sequences from strains included on a single sequencing flow cell. For each pair-wise comparison the length of the alignment achieved, expressed as the percentage of the total sequence length, was calculated and a distance matrix created. The heat map was created using the R statistical package and shows the clustered genomes determined by the default R heatmap function clustering methods ( http://www.r-project.org/). At the top of the figure, an indication of the relatedness between genomes is given. Mauve achieved pairwise genome sequence alignments of between 69.8 and 94.4% across our range of genomes. Strains are listed in the same order on the x and y axes; groupings discussed in the text are indicated along the top axis and the relevant strains are indicated by brackets on the right hand side axis, labelled with a Greek letter.