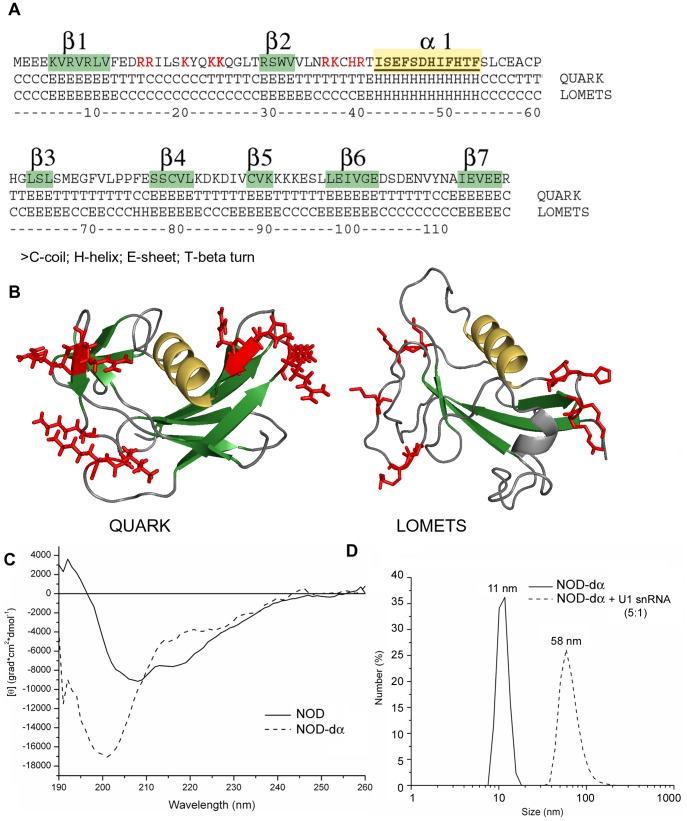
Figure 6. Prediction of NOD tertiary structure.
(A) NOD protein sequence annotated with the secondary structure elements predicted by the QUARK and LOMETS web-services. (B) The three-dimensional model of NOD was constructed using QUARK and LOMETS, with the positive amino acids which replacement with Alanines affect RNA-binding are shown in red letters (A) or in red sticks (B). β-sheets are shown in green, long α-helix is shown in yellow. The picture was prepared using PyMOL (www.pymol.org). (C) Circular dichroism far UV-light spectra of NOD and its mutant with the deleted α-helix (NOD-dα). Fluorescence intensity is given in relative units. (D) The hydrodynamic radius of the NOD-dα mutant as free protein particles or in complex with U1 snRNA, determined by DLS.