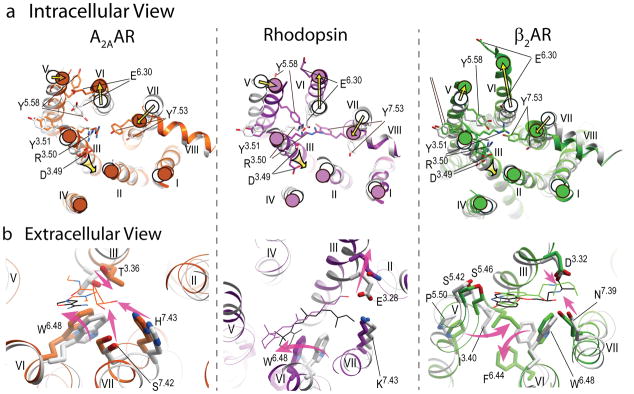
Figure 4.
Major conformational rearrangements and ligand-dependent triggers in the three available structural models of GPCR activation. a) Intracellular view: common shifts of the intracellular tips of transmembrane helices (yellow arrows), include outward swinging of helix VI, accompanied by movement of helix V, as well as inward shift of helix VII and axial shift of helix III. The established conserved microswitches, shown by stick presentations and labeled, undergo rotamer changes upon activation. b) Extracellular view: the key ligand-dependent “triggers” of GPCR activation (highlighted by the shaped magenta arrows) found in the orthosteric site. Note that trigger residues and their Ballesteros-Weinstein positions are not conserved between these receptors, and the directions of the helical shifts are different. Ligands are shown by thin lines with black carbons for all antagonists (ZM241385, 11-cis retinal and carazolol, respectively) and colored carbons for agonists (NECA, all-trans retinal and BI-167107 colored orange, purple and green, respectively). In all panels, inactive conformations are shown in gray, and corresponding activated conformations are colored orange for A2AAR, PDB codes 3EML (45) and 3QAK (19), purple for Rhodopsin, PDB codes 1GZM (114) and 2X72 (17), and green for β2AR, PDB codes 2RH1 (27) and 3SN6 (21).