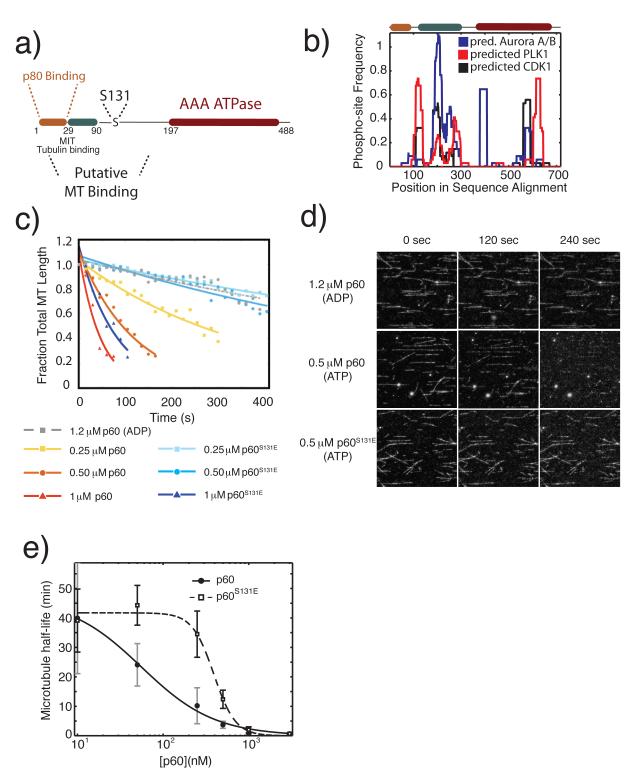
Figure 1. A phosphomimetic mutation in an N-terminal regulatory domain of p60 katanin directly inhibits microtubule severing.
a) Schematic domain structure of p60 katanin and the position of Serine 13124,25. b) The predicted domain regulatory potential for three mitotic kinases. Frequency of phospho-sites is calculated as the fraction of predicted phospho-sites within a sliding window of 30 amino acids across all sequences in the multiple sequence alignment. Predicted phospho-sites for CDK1, PLK1 and Aurora A and B combined were determined using the Group-based prediction system 2.1 online server26. Multiple sequence alignments were generated in Matlab (Mathworks) using the multialign function in the bioinformatics toolbox. c) Quantification of the time course of severing of taxol stabilized microtubules by purified X. laevis p60 katanin or p60S131E at indicated concentrations, measured in fraction of total microtubule length, and fit to an exponential decay model. Representative data from 3 independent experiments except 1.2 μM ADP, which was a single experiment to indicate the requirement for ATP. d) Images from the time lapse data quantified in c. Protein expression, purification, data collection, and processing were as previously described20. e) Plot of microtubule half-life relative to p60 concentration. Data were fitted to the 4 parameter logistic equation to indicate the variation in point of 50% maximal activity and identify the relative differences in Hill slope.