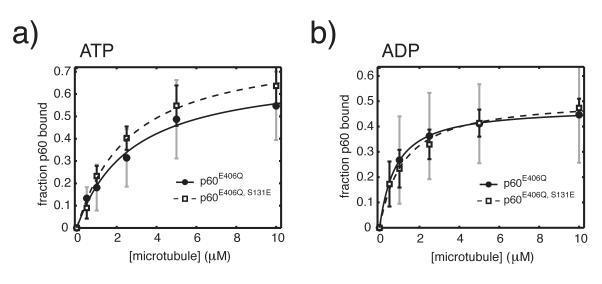
Figure 2. The affinity of p60 for microtubules is unchanged by a phosphomimetic mutation.
250 nM X. laevis p60 or p60S131E with either ATP (a) or ADP (b) was added to increasing concentrations of taxol-stabilized MTs from 0-10 μM in BRB80 with 10μM Taxol. Reactions were incubated at room temperature for 15 min, pelleted by centifugation and analyzed by SDS-PAGE. Binding data was fit to the quadratic binding equation as previously described20. Error in fitted parameters was estimated by monte carlo sampling within the experimentally determined error and fitting the resulting datasets27. This procedure generates hundreds of theoretical datasets that fit the measured data by randomly sampling a single value at each protein concentration that falls within the experimental error of that data point. The theoretical datasets are individually fitted normally. The maximum likelihood mean±SD was determined from 250 individual fits and provides the reported KD value and error. Plotted data is from two independent experiments with two separate preparations of each protein.