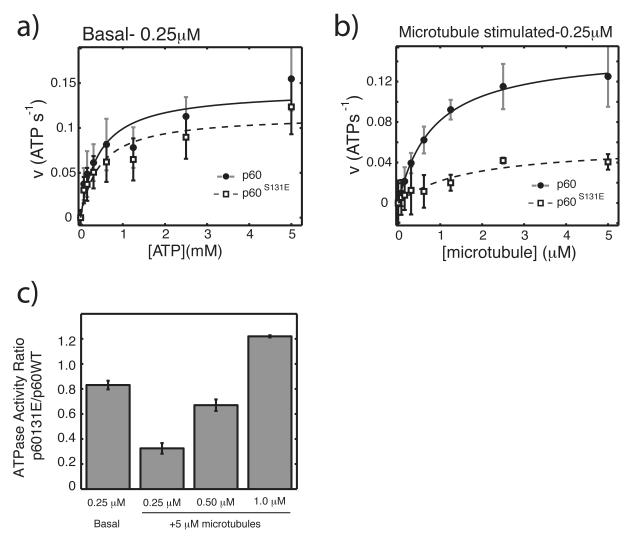
Figure 3. Microtubule stimulated ATPase activity is inhibited by a phosphomimetic mutation.
a) Measurement of basal ATPase activity of 250 nM p60 and p60S131E with increasing ATP concentration. b) Measurement of ATPase activity of 250 nM p60 and p60S131E in response to increasing microtubule concentration and 1mM ATP, corrected for basal activity. ATPase activity was determined using an enzyme-coupled assay as previously described except that reactions were in 20 mM HEPES pH 7.7, 25 mM potassium glutamate, 10% glycerol, 0.02% Triton-X100, 10 mM MgCl228. Data was fit to the Michaelis-Menton equation and estimates of the error in fitted parameters was determined by monte carlo sampling of data within the experimental error and fitting each resulting dataset27. The maximum likelihood mean±SD was then determined from 200 individual fits within the experimental error. c) ATPase activity of p60S131E relative to wild-type at 1mM ATP and 5 μM microtubules with propogated error. Data for basal and 250nM are the same as in part a and b, respectively. All data is from at least 3 independent experiments with two separate preparations of each protein.