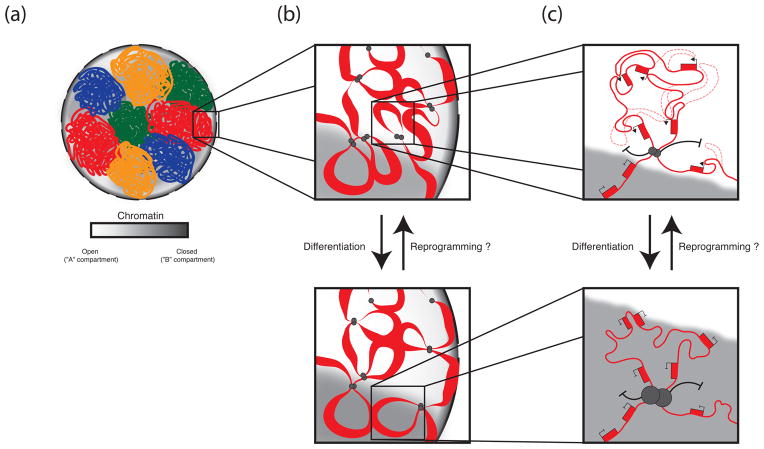
Figure 1. Hierarchical levels of genome organization and changes upon ESC differentiation.
Model of the 3D organization of the genome in ESCs and its changes during the course of differentiation. We infer this model by combining findings from many different cell types. (a) Chromosomes exist as discrete, minimally overlapping territories. At the megabase level, compartments of open (white) and closed (gray) chromatin coarsely divide the genome into regions enriched for features of euchromatin and those depleted of euchromatic features, respectively. Locus positioning within the open or closed compartment defines likely interaction partners, with loci in the open compartment interacting more frequently with other open loci, and those in closed more frequently with other closed loci. (b) Below the megabase, level the genome is divided into topological domains, which, we speculate, exist as large chromatin loops created by the juxtaposition of CTCF binding sites at TAD boundaries. These TADs function as modular units of genome organization whose member genes are often co-regulated, and we propose, localize as units within the nucleus and potentially switch between open and closed compartments during differentiation, somatic cell reprograming, or between different cell types as their euchromatic character changes. (c) Within TADs, enhancers and promoters loop together extensively and promiscuously to orchestrate cell type-specific gene expression profiles. Genes appear to be more likely co-regulated if they lie within the same TAD, as opposed to between TADs. During differentiation and transcription factor induced reprogramming, all genes within a TAD may switch their transcriptional state.