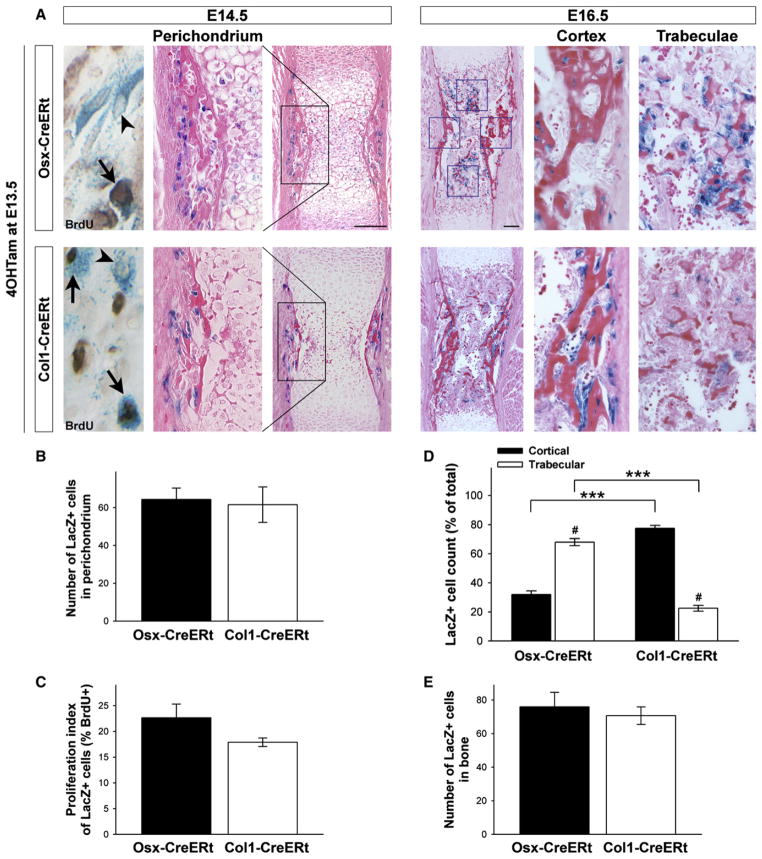
Figure 2. Perichondrial Osteoblast Lineage Cells Marked by Osx- or Col1-CreERt Expression Display Differential Fates in Developing Bones.
(A) Osx-CreERt (top) and Col1-CreERt (bottom) humerus sections of mice pulsed with 4OHTam at E13.5 and sacrificed at E14.5 (left) or at E16.5 (right) and stained for X-gal and eosin or BrdU (far left; arrowheads indicate single LacZ+ cells, arrows point at LacZ+/BrdU+ double-positive cells). Scale bars, 100 μm. (B and C) Quantification of the perichondrial labeling (B) and the proliferation index of the labeled cells (C) in Osx- and Col1-CreERt(Tg/−) mice at E14.5. The number of LacZ+ cells and the percentage that additionally stained positive for BrdU were determined in a defined region, encompassing the central portion of the perichondrium corresponding to the black boxed area in (A). Bars, mean ± SEM; n = 3.
(D and E) Distribution (D) and total number (E) of traced Osx/LacZ+ and Col1/LacZ+ cells at E16.5. Cells were counted in fixed cortical and trabecular bone areas (blue squares in A). ***p < 0.001 for comparison between genotypes; #p < 0.001 between locations. Bars, mean ± SEM; n = 5–9.
See also Figure S2 on the lineage tracing kinetics of the system.