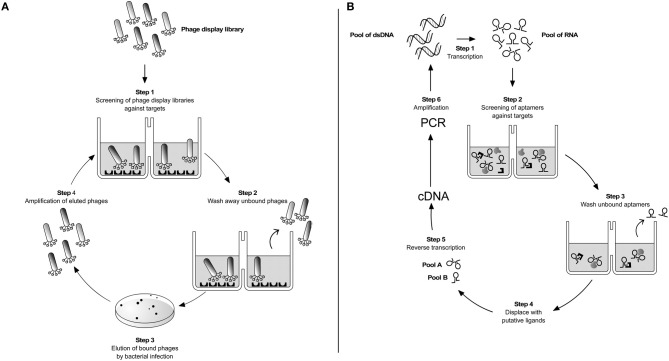
Figure 1.
Parasite targetting by combinatorial techniques. (A) Phage display. A phage library displaying potential ligand proteins on their surface is exposed to an immobilized target (step 1). After washing away unbound phages (step 2), binders are eluted by Escherichia coli infection and plated on LB-agar (step 3). Clones are then amplified producing a phage mixture that is enriched with relevant (i.e., binding) phage (step 4). The repeated cycling of these steps is referred as “panning”. At the end of 3–4 rounds of panning the enriched phage population is recovered by infection of a suitable bacterial host and sequenced to identify the interacting peptides or protein fragments. (B) SELEX. It is based on a stretch of single-stranded nucleic acid, which can be RNA or single-stranded DNA (ss-DNA). These are chemically synthesized to have a random stretch usually from 8 to 40 nucleotides, flanked by constant sequences. In the case of RNA SELEX, the synthetic DNA template is transcribed into a pool of 1013–1014 different RNA molecules (step 1). The pool is incubated with the desired targets and due to the sample diversity some of the aptamers will bind to their targets (step 2). After washing out unbound RNAs (step 3) the different RNA pools are displaced by incubation with ligands of interest (step 4). By reverse transcription (step 5) and PCR amplification (step 6) selected double-stranded DNAs are reconstructed. The same cycle is repeated over 8–12 times until purified sequences specific for a given ligand are selected. The DNAs are cloned and sequenced. This iterative method follows the same logic when single-stranded DNA sequences are used as aptamers instead of RNA (Ulrich and Wrenger, 2009).