Abstract
Background
Mouse limb bud is a prime model to study the regulatory interactions that control vertebrate organogenesis. Major aspects of limb bud development are controlled by feedback loops that define a self-regulatory signalling system. The SHH/GREM1/AER-FGF feedback loop forms the core of this signalling system that operates between the posterior mesenchymal organiser and the ectodermal signalling centre. The BMP antagonist Gremlin1 (GREM1) is a critical node in this system, whose dynamic expression is controlled by BMP, SHH, and FGF signalling and key to normal progression of limb bud development. Previous analysis identified a distant cis-regulatory landscape within the neighbouring Formin1 (Fmn1) locus that is required for Grem1 expression, reminiscent of the genomic landscapes controlling HoxD and Shh expression in limb buds.
Results
Three highly conserved regions (HMCO1-3) were identified within the previously defined critical genomic region and tested for their ability to regulate Grem1 expression in mouse limb buds. Using a combination of BAC and conventional transgenic approaches, a 9 kb region located ~70 kb downstream of the Grem1 transcription unit was identified. This region, termed Grem1 Regulatory Sequence 1 (GRS1), is able to recapitulate major aspects of Grem1 expression, as it drives expression of a LacZ reporter into the posterior and, to a lesser extent, in the distal-anterior mesenchyme. Crossing the GRS1 transgene into embryos with alterations in the SHH and BMP pathways established that GRS1 depends on SHH and is modulated by BMP signalling, i.e. integrates inputs from these pathways. Chromatin immunoprecipitation revealed interaction of endogenous GLI3 proteins with the core cis-regulatory elements in the GRS1 region. As GLI3 is a mediator of SHH signal transduction, these results indicated that SHH directly controls Grem1 expression through the GRS1 region. Finally, all cis-regulatory regions within the Grem1 genomic landscape locate to the DNAse I hypersensitive sites identified in this genomic region by the ENCODE consortium.
Conclusions
This study establishes that distant cis-regulatory regions scattered through a larger genomic landscape control the highly dynamic expression of Grem1, which is key to normal progression of mouse limb bud development.
Background
During embryonic development, the spatio-temporal expression of transcriptional regulators and morphogenetic signals is tightly controlled. In fact, different types of congenital malformations are caused by mutations affecting specific cis-regulatory and non-coding genomic regions that alter the expression of genes or gene clusters in specific tissues (reviewed in ref. [1-3]). Genome-wide functional mapping revealed many of the large genomic landscapes that control the expression of developmental regulator genes in a global and tissue-specific manner [4]. This study and others reveal that the cis-regulatory regions controlling gene expression in a particular tissue are often located several hundred kilobases (kb) or even megabases (Mb) up- or downstream of the transcriptional start sites in other loci, and may control the expression of several genes [5-7]. Recently, it has been shown how multiple cis-regulatory regions interact to control the expression of the 5’HoxD gene complex in the presumptive digit territory. The transcriptionally active part of the 5’HoxD gene cluster forms a so-called regulatory archipelago in which dispersed cis-regulatory elements co-operate to control gene expression in the distal limb bud by interacting with proximal promoters over large distances [8]. In fact, regulatory landscapes with distant and dispersed cis-regulatory regions seem to be a recurring theme in the dynamic spatio-temporal regulation of genes that are essential for limb bud development (reviewed in ref. [2,9,10], see also below).
Vertebrate limb bud development is controlled by interactions between two main signalling centres, the apical ectodermal ridge (AER) and the zone of polarizing activity (ZPA) located in the posterior limb mesenchyme. These two signalling centres interact as part of a self-regulatory feedback signalling system involving several signalling pathways [11]. In particular, ZPA cells produce the Sonic Hedgehog (SHH) signal, which together with GREMLIN1-mediated antagonism of Bone Morphogenetic Proteins (BMPs) in the posterior-distal limb bud mesenchyme propagates Fibroblast Growth Factor (FGF) signalling in the AER [11-14]. This SHH/GREM1/AER-FGF feedback-signalling loop promotes distal progression of limb bud outgrowth and formation of the autopod which gives rise to carpals and digit rays (reviewed in ref. [15]). SHH expression in the posterior limb bud mesenchyme is controlled by a cis-regulatory region located about 800 kb upstream of the transcriptional start site. Deletion of this ZPA regulatory region (ZRS) results in loss of distal limb structures [16], while point mutations within the ZRS result in anterior ectopic Shh expression and duplications of the thumb and/or anterior digits in different species [17,18]. In cells actively transcribing Shh, the ZRS loops out of its chromosomal territory to the Shh transcription unit, which reveals how distant cis-regulatory elements control gene expression [19]. Recently, several transcriptional regulators have been identified that control Shh expression by directly interacting with the distant ZRS region. These include HOX proteins, the bHLH transcription factor HAND2, and ETS transcription factors, providing a glimpse of the complex transcriptional regulation of Shh in the posterior limb bud mesenchyme [20-22].
SHH signalling is transduced by the GLI family of transcriptional regulators and inhibits the constitutive processing of the full-length GLI3 activator to its repressor form GLI3R (see e.g. ref. [23]). Of the three GLI transcription factors expressed during limb bud development, only Gli3 is essential on its own (see e.g. ref. [24]). The inactivation of Gli3 alters morphogenetic feedback signalling and results in formation of additional anterior digits [23,25-27]. In particular, GLI3 restrains the proliferation of mesenchymal progenitors in the anterior limb bud mesenchyme and promotes initiation of digit ray chondrogenesis by directly repressing Grem1 expression during handplate formation [26].
We have previously shown that the expression of Grem1 in the limb bud mesenchyme is controlled by a large genomic landscape downstream of its transcription unit [7]. In fact, several of the classical limb deformity mutations that disrupt distal limb bud development and formation of metanephric kidneys in the mouse are caused by deletions affecting this cis-regulatory landscape rather than directly altering the Grem1 gene products [7,13,28]. Molecular and genetic analysis in the mouse identified a 70 kb genomic region located downstream of the Grem1 transcription unit within the neighbouring Formin1 (Fmn1) gene that is required for Grem1 expression in the limb bud mesenchyme [7,29]. Detailed analysis of this Grem1 genomic landscape revealed similarities with the global control region (GCR) that controls the expression of 5’HoxD genes in the limb bud mesenchyme [6], but did not reveal the structural nature and transacting factors and/or signalling pathways controlling these cis-regulatory regions.
To gain further insights into the Grem1 landscape, the potential cis-regulatory activities of the three highest evolutionarily conserved genomic regions within the 70 kb Grem1 critical region were analysed. In addition to its expression in the posterior mesenchyme, emphasis was also given to the coordinated distal-anterior expansion of Grem1 expression, which is key to orderly progression of limb bud development [14,30]. Combining BAC with conventional transgenic approaches, we identified a 9 kb genomic region that is able to recapitulate major but not all aspects of the dynamic spatio-temporal Grem1 expression in the limb bud mesenchyme. This 9 kb region was termed Grem1 Regulatory Sequence 1 (GRS1), and contains a core sequence that is essential to express a LacZ transgene under control of a ß-globin minimal promoter in a Grem1-like pattern. The GRS1 region drives gene expression into the posterior and subsequently distal-anterior mesenchyme, i.e. reproduces aspects of the distal-anterior expansion of Grem1 expression during progression of limb bud development. By crossing GRS1 transgenic mice into different mutant contexts, we establish that the GRS1 region is controlled by inputs from both the SHH and BMP signalling pathways in limb buds. In addition, chromatin immunoprecipitation (ChIP) shows that GLI3 interacts with the conserved HMCO1 sequences in the GRS1 region. The functionally relevant cis-regulatory regions identified by the present and two previous studies [31,32] map to the endogenous DNAse I hypersensitive sites within the Grem1 genomic landscape recently identified by the ENCODE consortium [33,34]. This indicates that the Grem1 cis-regulatory landscape is composed of at least five active cis-regulatory regions that control the spatio-temporal expression of Grem1 in mouse embryos and limb buds.
Results and Discussion
Identification of conserved limb bud regulatory regions within the Grem1 genomic landscape
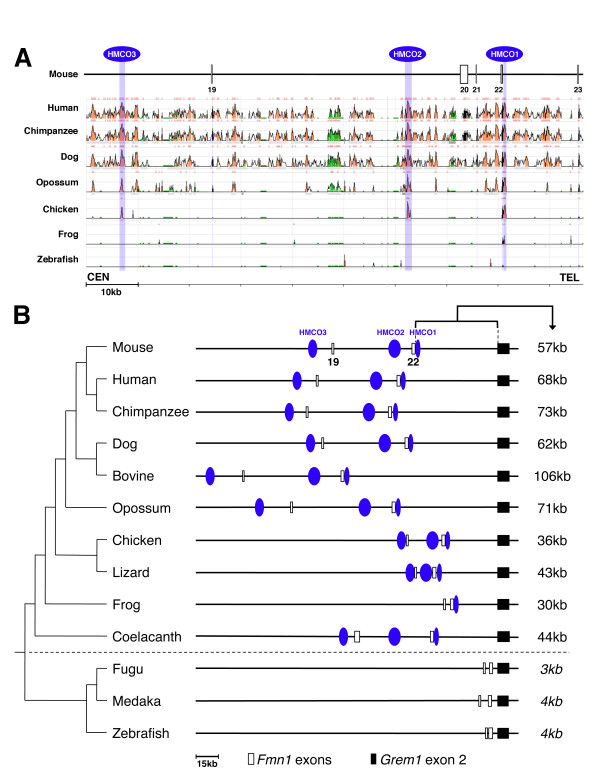
Using functional genetics in the mouse, we previously identified a ~70 kb region located downstream of Grem1 that is required for its expression in the limb bud mesenchyme (Figure 1A, [7]). This limb bud cis-regulatory region is located between coding exons 19 and 23 of the neighbouring Fmn1 gene. The Evolutionary Conserved Regions (ECR) genome browser ( http://ecrbrowser.dcode.org/) was used for multiple sequence alignments to compare the mouse genome with different mammalian and the chicken genomes. This analysis revealed several blocks of highly conserved sequences among mammalian species, but only three of them were also highly conserved in the chicken and termed HMCO1, 2 and 3 (Human-Mouse-Chicken-Opossum conserved sequences 1 to 3, Figure 1A). The most conserved parts of these three HMCO core regions are ~80% identical (for details see Additional file 1 and Additional file 2). Of these three regions only HMCO1 is also present in the orthologous genomic region in frogs (Figure 1A), which express Grem1 during limb bud development [35]. Despite the fact that Grem1 is expressed during fin bud development [36], no HMCO homologies are present in the zebrafish genome (Figure 1A). ClustalW2 alignments ( http://www.ebi.ac.uk/Tools/msa/clustalw2/) of the Fmn1Grem1 regions from different species revealed the absence of HMCO homologies in other ray-finned fish species such as fugu and medaka (Figure 1B). In contrast, all three HMCO regions are present in the genome of coelacanth, a lobe-finned fish closely related to tetrapods (Figure 1B and Additional file 1) [37]. As the Grem1 and Fmn1 loci are linked in all species analysed (Figure 1B), the distance between the orthologous Fmn1 coding exon 22 (located adjacent to HMCO1) and Grem1 coding exon 2 was determined. During evolution, this distance increased as it is smallest in the genomes of ray finned fishes and at least ~10-fold larger in tetrapods and coelacanth (Figure 1B). In addition, the distance between Fmn1 exon 19 (close to HMCO3) and exon 22 correlates well with the presence or absence of the three HMCO regions (Figure 1B and data not shown). In ray-finned fishes and frogs (lacking all or specifically HMCO2, 3, respectively), the distance is ~3-7 kb, while in mammals and coelacanth it ranges from ~50-70 kb. Thus, the increase of intronic and intergenic regions in the Fmn1-Grem1 landscape correlates well with appearance of the three HMCO regions during vertebrate evolution.
Figure 1.
Identification of three highly conserved non-coding regions in tetrapods and a lobe-finned fish. (A) Sequence alignment of the genomic region critical for Grem1 expression in mouse limb buds [7] using the ECR browser with the mouse genome release 9 (mm9) as reference genome. The critical genomic region on mouse chromosome 2 is shown in centromeric (CEN) to telomeric (TEL) orientation and is located downstream of the Grem1 coding exons. As the critical genomic region is part of the Fmn1 locus, the interspersed Fmn1 coding exons are indicated as open boxes in the scheme. Three blocks of highly conserved non-coding sequences were identified (HMCO1-3) and are indicated in blue. Conserved coding regions are indicated in black and non-coding regions conserved ≥74% over ≥100 bp are coloured salmon. The peak detected in the region of HMCO1 in the zebrafish genome corresponds to Fmn1 exon 22. Regions consisting of repetitive sequences are shown in green (see Additional file 2). (B) Conserved linkage between the Grem1 and Fmn1 loci in vertebrates. Increased intergenic distances correlate with the presence of HMCO regions in tetrapods and coelacanth in contrast to ray-finned fishes. The phylogenetic tree analysis was done with the UCSC multiple alignment functions to align the 3' part of the Fmn1 locus and the Grem1 locus from different species. Open boxes represent the orthologous Fmn1 coding exons 19 and 22, black box represents Grem1 coding exon 2. The intergenic distances between Fmn1 orthologous exon 22 and Grem1 coding exon 2 are indicated to the right of the scheme. ENSEMBL genomes used for alignment: mouse: M. musculus (mm10); human: H. sapiens (hg19); chimpanzee: P. troglodytes (panTro3); dog: C. familiaris (canFam2); bovine: B. taurus (bosTau6); opossum: M. domestica (monDom5); chicken: G. gallus (galGal3); lizard: A. carolinensis (anoCar2); frog: X. tropicalis (xenTro2); coelacanth: L. chalumnae (LatCha1); fugu: T. rubripes (fr3); medaka: O. latipes (oryLat2); zebrafish: D. rerio (danRer7).
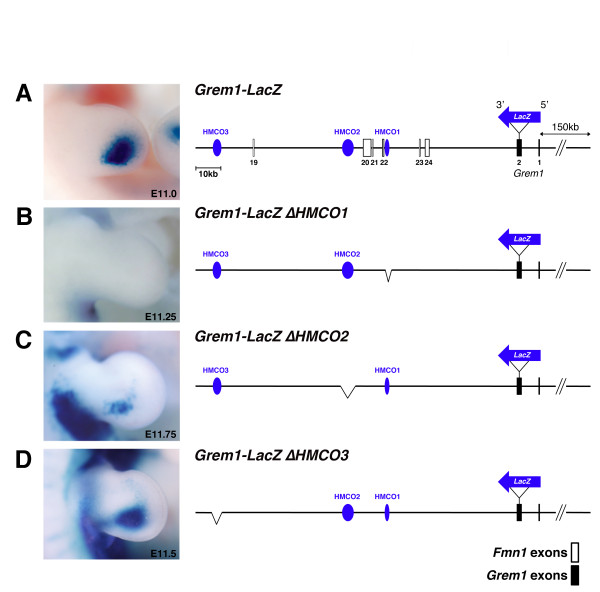
To determine the requirement of each of these three HMCO regions for Grem1 expression in mouse limb buds, we used a BAC-based strategy in combination with analysis of transgenic founder embryos (Figure 2A-D and Additional file 3). A 250 kb mouse genomic BAC encoding the critical region and the Grem1 transcription unit was used to fuse the LacZ gene in frame with the Grem1 ORF [7]. We assessed the expression of the LacZ reporter transgene by analysing the spatio-temporal distribution of ß-galactosidase activity (Figure 2A-D). Expression of the Grem1-LacZ fusion protein in the forelimb bud mesenchyme was detected in a posterior-distal domain (Figure 2A), mimicking the early endogenous Grem1 expression rather accurately. Therefore, the Grem1-LacZ BAC construct was used to engineer deletions of the three HMCO regions and determine their requirement for Grem1-LacZ expression in the posterior mesenchyme. To control for reproducible generation of expressing BAC transgenic founder embryos, the Grem1-LacZ BAC (Figure 2A) was injected in parallel to the BACs with engineered deletions (Figure 2B-D). While the control Grem1-LacZ BAC was always robustly expressed in the posterior mesenchyme (Figure 2A), deletion of the 520 bp HMCO1 core region resulted in complete loss of LacZ expression from limb buds (Figure 2B, n = 5/7 embryos with LacZ expression, for details see Additional file 3). In contrast, deletion of the 1279 bp HMCO2 core region only caused partial loss of LacZ expression from the posterior limb bud mesenchyme (Figure 2C; Additional file 3). LacZ remained expressed normally in the majority of founder embryos carrying a 924 bp deletion of the HMCO3 region (Figure 2 = 3/5 embryos with LacZ expression, for details see Additional file 3). Taken together, this BAC transgenic analysis establishes HMCO1 as most critical for Grem1 expression in the posterior-distal limb bud mesenchyme. As the HMCO1 core region is highly conserved in tetrapods and lobe-finned fish but not ray-finned fish (Figure 1B), it likely represents a cis-regulatory region important to tetrapod evolution. The other two HMCO regions might contribute to robust expression of Grem1LacZ in the posterior mesenchyme, as in particular the deletion of HMCO2 results in significantly reduced LacZ expression (Figure 2C).
Figure 2.
A BAC-based strategy to assess the requirement of highly conserved non-coding regions (HMCO1-3) for Grem1 expression in mouse limb buds. (A) A 250 kb BAC spanning the genomic region from Fmn1 exon 19 to 150 kb up-stream of the Grem1 transcription is able to drive expression of a LacZ transgene (blue arrow; inserted in-frame into the Grem1 ORF) into the posterior limb bud mesenchyme in transgenic founder embryos. (B) Deletion of the HMCO1 core region (520 bp) abolishes the LacZ expression. (C, D) Deletion of the core HMCO2 region (1279 bp) results in reduced LacZ expression, while deletion of HMCO3 (924 bp) does not alter the LacZ distribution. ß-galactosidase activity colours expressing cells blue.
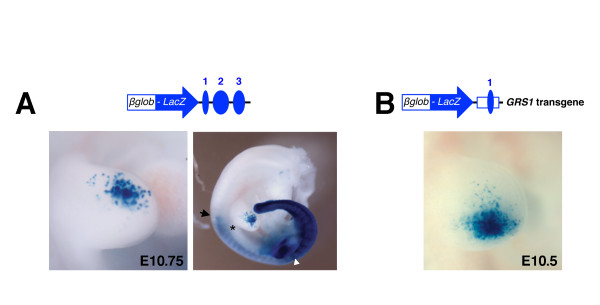
In an attempt to gain further insight into the enhancer potential and possible interactions of HMCO2/3 with HMCO1, conventional transgenic approaches using a minimal human ß-globin promoter (ßglob-LacZ) were employed. However, neither individual HMCO regions (data not shown) nor in a combination of all three was able to drive robust expression of the ßglob-LacZ transgene in the posterior limb bud mesenchyme (Figure 3A, compare to Figure 2A). Expression of the ßglob-LacZ transgene under control of all three HMCO core regions resulted in scattered LacZ positive cells in the anterior-distal mesenchyme of forelimb buds (left panel, Figure 3A, n = 3/3). In contrast, the transgene was strongly expressed in the posterior embryo including hindlimb buds (right panel, Figure 3A). These results show that a transgene consisting of an array of the HMCO core regions is unable to drive LacZ expression into the posterior forelimb bud mesenchyme. This indicates that additional elements of the Grem1 genomic landscape are required for Grem1 expression in limb buds.
Figure 3.
A 9 kb GRS1 transgene encompassing HMCO1 drives ßglob-LacZ expression into the posterior limb bud mesenchyme. (A) A transgene encoding the three HMCO core regions downstream of the ßglob-LacZ minimal promoter and reporter results in aberrant ß-galactosidase activity in the anterior-distal limb bud mesenchyme. An asterisk marks the posterior border of the forelimb bud. A black arrow marks the anterior border of LacZ expression in the trunk. A white arrowhead points to the hindlimb bud. (B) The GRS1-ßglob-LacZ transgene is expressed in the posterior limb bud mesenchyme of transgenic founder embryos. The 9 kb GRS1 region was inserted downstream of the ßglob-LacZ reporter to keep the same arrangement as in the endogenous Grem1 locus (Figure 2A).
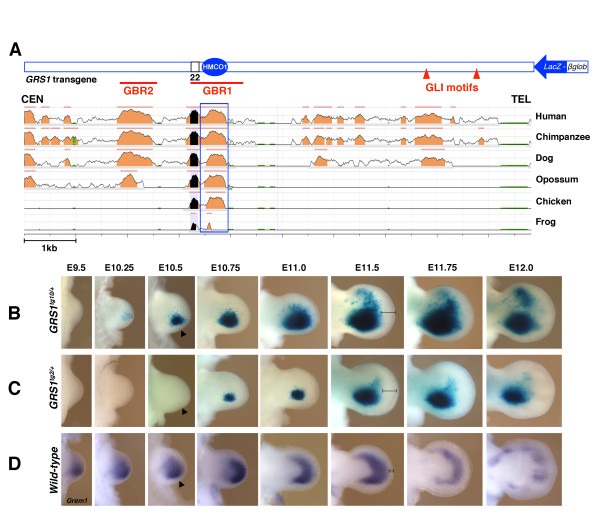
Next, we assessed to which extent a larger genomic fragment containing HMCO1 and flanking regions could drive ßglob-LacZ expression into the posterior limb bud mesenchyme (Figure 3B). Analysis of transgenic founder embryos revealed that this transgenic construct drives robust ßglob-LacZ expression in the posterior limb bud mesenchyme (Figure 3B, see also Figure 4), strikingly similar to the domain of the parental Grem1-LacZ BAC construct (Figure 2A). Taken together, these results indicate that this cis-regulatory region enhances Grem1 expression in the posterior limb bud mesenchyme, which prompted us to term this 9 kb region Grem1 Regulatory Sequence 1 (GRS1, Figure 4A). GRS1 encompasses the HMCO1 region next to Fmn1 coding exon 22 and a further downstream region that is highly conserved in mammals. This region, like HMCO1 overlaps with a previously identified GLI binding region (GBR, [32]). In addition, two potential GLI binding sites were identified by in silico analysis (Figure 4A).
Figure 4.
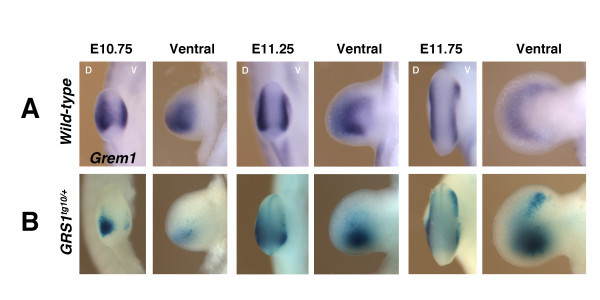
Comparison of GRS1 -dependent ß-galactosidase activity with endogenous Grem1 transcripts during limb bud development. (A) ECR browser alignment of the 9 kb GRS1 region containing HMCO1. The mouse genome (mm9) is used as reference genome. The conservation threshold was set ≥74% identity over ≥100 bp (see legend to Figure 1A for details). Above the sequence alignment, a schematic representation of the GRS1-ßglob-LacZ transgene is shown. GBR: GLI binding region [32]; GLI motifs: consensus GLI binding sites identified by in silico analysis. (BC) Distribution of ß-galactosidase activity in forelimb buds of heterozygous embryos of the tg10 (10 copies, GRS1tg10/+, B) and the tg2 (2 copies, GRS1tg2/+, C) mouse strains between embryonic days E9.5 to E12. (D) Distribution of endogenous Grem1 transcript in the corresponding stages of wild-type forelimb buds. Arrowheads at E10.5 point to the ß-galactosidase activity and Grem1 transcript in the posterior limb bud mesenchyme. The brackets at E11.5 serve to indicate the distance between the ß-galactosidase activity (BC) or Grem1 expression domain (D) and the AER. Note that ß-galactosidase activity fails to expand distally in comparison to the Grem1 domain.
Spatio-temporal activity of the GRS1 region during mouse limb bud development
To analyze comparatively the activity of the GRS1 region with respect to the spatio-temporal regulation of Grem1 expression, transgenic mouse strains expressing the GRS1-ßglob-LacZ reporter construct were established (Figure 4). Seven transgenic founders were obtained. In three independent strains, LacZ was expressed in the posterior fore- and hindlimb bud mesenchyme in a pattern comparable to the transgenic founder embryos (Figure 3B and data not shown). Two of these three transgenic strains were analyzed in detail and used to study the spatio-temporal LacZ distribution (Figure 4B, C and Additional file 4). Initial analysis revealed that ß-galactosidase activity of the LacZ transgene was detected earlier in one strain and is significantly higher than in the other strain (Figure 4B, compare to Figure 4C). As levels did not change during subsequent generations (data not shown), this is likely due to differences in transgene copy number and/or integration site. Therefore, the copy number of both strains was determined by real-time qPCR analysis (Additional file 4). This analysis revealed that ten copies of the GRS1-ßglob-LacZ transgene were integrated into the genome of the strain expressing higher LacZ levels (GRS1tg10), while only two copies were detected in the GRS1tg2 strain (Additional file 4). In GRS1tg10/+ embryos, ß-galactosidase activity is first detected at ~ E10.25 in the posterior forelimb bud mesenchyme and continuously increases until ~ E11.75 (Figure 4B). From ~ E11.0 onwards, scattered positive cells were detected in the anterior mesenchyme, and this anterior expression increased during distal limb bud outgrowth to form a crescent in the distal-anterior autopod (right panels, Figure 4B). By E11.75, ß-galactosidase activity was rather variable, such that the anterior crescent separated from the posterior domain in some forelimb buds. In GRS1tg2/+ embryos, the spatio-temporal pattern is similar, but expression levels are significantly reduced due to the lower transgene copy number, resulting in detection of ß-galactosidase activity from only ~ E10.5 onwards (Figure 4C, arrowhead). In both strains, GRS1 drove expression of the ßglob-LacZ reporter specifically in the limb bud mesenchyme, despite some low ß-galactosidase activity detected in the developing eyes of GRS1tg10/+ transgenic embryos (Additional file 4). This analysis reveals the robust nature of the GRS1 region, which functions in positive regulation of Grem1 expression in the limb bud mesenchyme. The posterior domain of ß-galactosidase activity at E10.5 (Figure 4B) is comparable to the Grem1 transcript distribution in wild-type limb buds at earlier stages (E9.5-E10.25, Figure 4D). This temporal delay is likely due to postponed transcriptional activation and/or up-regulation of the GRS1 transgene as the establishment of a posterior LacZ expression domain is also only apparent at E10.5 in GRS1tg10/+ transgenic limb buds (Additional file 5). Therefore, additional cis-regulatory regions likely control the temporally correct early onset of Grem1 expression. Similar delays in the onset of LacZ reporter gene expression have been previously observed by analyzing the ZRS cis-regulatory region that controls Shh expression in mouse limb buds [38]. During distal progression of limb bud development, endogenous Grem1 expression expands distal-anterior within the developing handplate and begins to fade by E11.75 (Figure 4D), due to termination of the SHH/GREM1/AER-FGF feedback loop and GLI3-mediated repression in the anterior limb bud mesenchyme [11,26,39,40]. In contrast, ß-galactosidase activity remains high in the posterior limb bud mesenchyme of GRS1 transgenic embryos and the distal-anterior expansion of its expression is significantly delayed and only occurs as transcription of the endogenous Grem1 locus is starting to terminate (E11.5 onwards Figure 4B, C, compare to Figure 4D).
Grem1 expression is restricted to dorsal and ventral limb bud mesenchyme and excluded from the chondrogenic core mesenchyme (Figure 5A), which is relevant to BMP-mediated induction of chondrogenesis (ref. [26,41] and Benazet et al., submitted). Furthermore, the LIM-homeodomain transcription factors Lhx2 and Lhx9 have been implicated in regulating Grem1 expression predominantly in the ventral limb bud mesenchyme in response to SHH signalling [42]. Therefore, the extent to which the dorso-ventral transcript distribution is maintained by the GRS1 transgene was assessed (Figure 5B). Indeed, the expression of both the high (Figure 5B) and low copy transgenes (Additional file 6) remained excluded from the core mesenchyme throughout limb bud outgrowth and patterning. Similar to Grem1 transcripts, ß-galactosidase activity is higher dorsally than ventrally (Figure 5B, compare to Figure 5A). By E11.75, ß-galactosidase activity expands anteriorly in both the dorsal and ventral mesenchyme (E11.75, Figure 5B), reminiscent of the crescent of Grem1 transcript (Figure 5A and see before), which indicates that the GRS1 contains cis-regulatory regions controlled by Lhx transcription factors [42]. Taken together, the results shown in Figures 4 and 5 establish that the 9 kb GRS1 cis-regulatory region is able to recapitulate major aspects of Grem1 expression in the limb bud mesenchyme. In particular, the GRS1 also recapitulates aspects of the distal-anterior expansion of Grem1 expression in limb buds (Figure 45), which was not observed in previous attempts to identify cis-regulatory regions that control Grem1 expression in limb buds [7,32]. The fact that the GRS1 transgene recapitulates some aspects of this distal-anterior expansion is important, as Grem1 transcription normally expands anteriorly in register with posterior AER-FGFs and allows propagation of SHH/GREM1/AER-FGF signalling [14,30]. However, the delay in activation and lack of termination of the GRS1 transgene (Figure 4) indicates that other regulatory inputs from the Grem1 genomic landscape are required to regulate its dynamic expression in mouse limb buds.
Figure 5.
Dorsal and ventral restriction of GRS1-mediated ß-galactosidase activity. (A) Grem1 transcript remains restricted to the dorsal and ventral forelimb bud mesenchyme in wild-type embryos throughout limb bud development. (B) GRS1-mediated expression of ßglob-LacZ is able to correctly restrict ß-galactosidase activity along the dorso-ventral limb bud axis. D: dorsal; V: ventral.
The GRS1 cis-regulatory region integrates inputs from the SHH and BMP signalling pathways
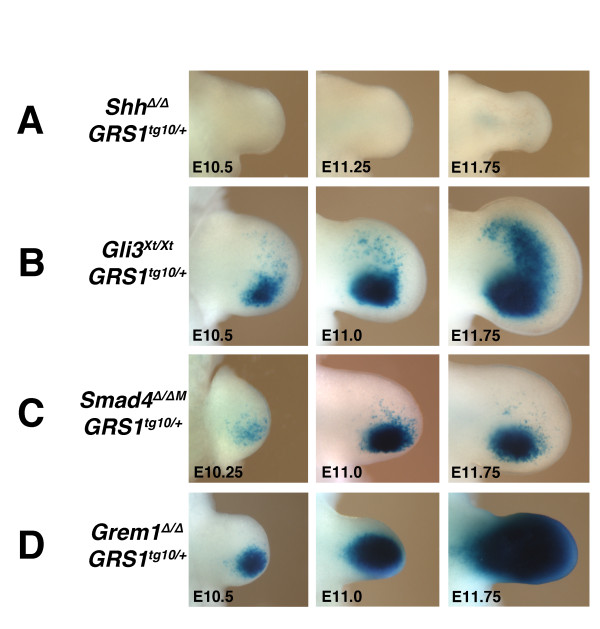
During limb bud initiation, mesenchymal BMP4 signal transduction is likely required to activate Grem1 expression in the posterior mesenchyme, while SHH is primarily required for up-regulation and distal-anterior expansion during progression of limb bud development [11,14,43]. Furthermore, GLI3 in the anterior mesenchyme and AER-FGF signal transduction are required to restrict and eventually terminate Grem1 expression (starting ~ E11.5-11.75) [26,40]. Therefore, GRS1tg10/+ embryos lacking key components of both the SHH and BMP signalling pathway in their limb buds were generated to gain insight into the possible direct impact of these main signalling pathways on the GRS1 element (Figure 6). Analysis of ShhΔ/ΔGRS1tg10/+ embryos revealed the complete absence of ß-galactosidase activity in Shh-deficient forelimb buds (Figure 6A). This contrasts with the endogenous Grem1 expression, which is activated but not maintained in Shh-deficient limb buds [11,14]. These results show that activation of the GRS1 region depends on SHH signalling, which indicated that it could also participate in ectopic Grem1 activation due to anterior ectopic SHH signalling in mouse [14] and chicken limb buds [12]. Furthermore, GLI3 proteins interact with specific cis-regulatory regions in the Grem1 genomic landscape [32] and genetic analysis has shown that Gli3 is essential for the spatio-temporally controlled restriction and subsequent termination of Grem1 expression in the distal anterior mesenchyme [25,26,44]. In Gli3-deficient GRS1 transgenic (Gli3Xt/XtGRS1tg10/+) forelimb buds, ß-galactosidase activity in the posterior limb buds is comparable to GRS1tg10/+ limb buds, while expression in the anterior mesenchyme is significantly increased by E11.75 (Figure 6B, compare to right-most panels, Figure 4B). This late up-regulation of anterior ß-galactosidase activity indicates that GRS1 is required for GLI3-mediated termination of Grem1 expression in the anterior limb bud mesenchyme, which is essential for the spatio-temporally correct initiation of mesenchymal condensations and chondrogenic differentiation of anterior digits [26].
Figure 6.
The GRS1 transgene is controlled by both SHH and BMP activity. (A) GRS1-dependent ß-galactosidase activity is completely lost from Shh-deficient limb buds (ShhΔ/ΔGRS1tg10/+). (B) In contrast, inactivation of Gli3 results in anterior up-regulation of ß-galactosidase activity at E11.75 (Gli3Xt/XtGRS1tg10/+). (C) ß-galactosidase activity fails to expand anteriorly in Smad4-deficient limb buds (Smad4Δ/ΔMGRS1tg10/+). (D) ß-galactosidase activity is up-regulated in Grem1-deficient limb buds from E11.0 onward (Grem1Δ/ΔGRS1tg10/+).
BMPs control Grem1 activation in the posterior limb bud mesenchyme and directly modulate its expression as part of the self-regulatory SHH/GREM1/AER-FGF feedback signalling system. Therefore, a conditional Smad4 loss-of-function allele [45] in combination with the Prx1-Cre recombinase strain [46] was used to inactivate Smad4 (Smad4Δ/ΔM) and thereby canonical BMP signal transduction in the limb bud mesenchyme. In Smad4Δ/ΔM mutant limb buds, endogenous Grem1 expression is down-regulated but not lost, while Shh expression remains (Benazet et al., submitted). In Smad4Δ/ΔMGRS1tg10/+ forelimb buds, ß-galactosidase activity appears normal in the posterior mesenchyme, while distal-anterior expansion fails to occur (Figure 6C, compare to Figure 4B). This indicates that GRS1 activity is modulated by SMAD4-mediated signal transduction during the progression of limb bud development. These results indicate that also the distal-anterior expansion of Grem1 expression depends on BMP activity and agree with the observation that genetic lowering of Bmp4 results in a global reduction of Grem1 expression in the limb bud mesenchyme [11].
Finally, in Grem1-deficient embryos, BMP activity is increased due to reduced BMP antagonism, which results in up-regulation of non-functional Grem1 transcript [13]. As BMP signal transduction modulates GRS1 activity during advanced limb bud development (Figure 6C), we also determined the potential influence of Grem1 deficiency on GRS1 activity. In Grem1Δ/ΔGRS1tg10/+ E10.5 limb buds ß-galactosidase activity is initially similar to GRS1tg10/+ limb buds (Figure 6D, compare to Figure 4B). Subsequently, ß-galactosidase activity is increased and by E11.75 the entire Grem1Δ/ΔGRS1tg10/+ limb bud is positive (Figure 6D). These alterations of ß-galactosidase activity in Smad4Δ/ΔM and Grem1Δ/Δ forelimb buds reveal that GRS1 activity is extensively modulated by changes in BMP activity, in particular also the anterior expansion of its expression. Taken together, this genetic analysis (Figure 6) shows that GRS1 activity critically depends on SHH and is modulated extensively by BMP signal transduction. This analysis identifies the GRS1, located ~70 kb downstream of the Grem1 transcription unit as a cis-regulatory region that integrates inputs by both SHH and BMP signal transduction. However, as previous analysis provided good evidence that Grem1 is activated by mesenchymal BMP signalling upstream of establishing the SHH/FGF feedback loop [11], these studies point to the existence of additional unknown BMP response regions that activate Grem1 expression in the posterior limb bud mesenchyme.
Endogenous GLI3 proteins are part of the chromatin complexes interacting with specific parts of the GRS1 region in mouse limb buds
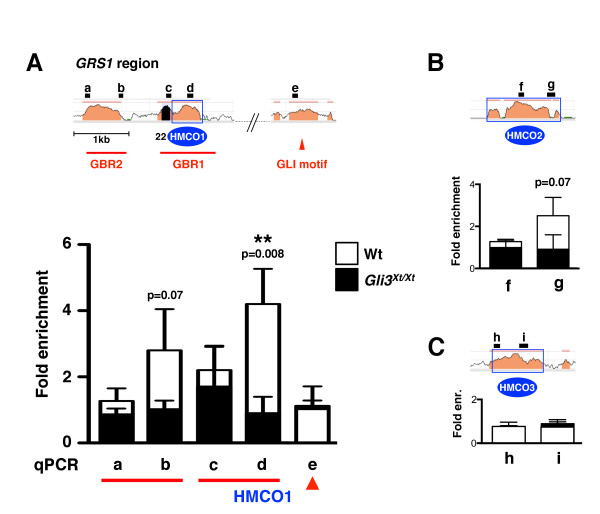
The observed loss of GRS1 activity in Shh-deficient limb buds (Figure 6A) indicated that the GRS1 region could be regulated directly by GLI proteins. Indeed, two GLI binding regions without consensus GLI binding motif (GBR1 and GBR2, [32]) were previously mapped to the 9 kb GRS1 region using transgene-mediated expression of an epitope-tagged GLI3 transgene in combination with chromatin immunoprecipitation (ChIP). To study the potential interaction of endogenous GLI3 proteins with the GRS1 region in its normal genomic context in wild-type limb buds, ChIP analyses using antibodies that detect both the GLI3 activator and repressor protein isoforms were performed [26,47]. GLI3 ChIP using extracts of wild-type and Gli3-deficient limb buds at E11.5 revealed significant enrichment of one specific region within GRS1 by real-time qPCR analysis (Figure 7A). A significant ~4-fold enrichment of an amplicon located within HMCO1 was observed by comparing wild-type to Gli3-deficient limb buds (amplicon “d”; p = 0.008) and to a less conserved region (amplicon “e”; located outside HMCO1 close to a putative GLI binding site; Figure 7A). In contrast, no significant enrichments of the other amplicons located in GRS1 were detected (Figure 7A). As the amplicons located in HMCO2 (Figure 7B) and HMCO3 (Figure 7C) were also not significantly enriched, the GLI3-containing chromatin complexes appear to interact rather specifically with a region within the highly conserved HMCO1 in wild-type limb buds. These results not only corroborates the identity of previously identified GBR1 [32], but indicate that this region mediates the effects of SHH signal transduction on GRS1 activity in the posterior limb bud mesenchyme.
Figure 7.
Endogenous GLI3 complexes bind to specific HMCO and GBR regions in the Grem1 genomic landscape. (A) Several amplicons were designed to detect the conserved and functionally relevant regions within the GRS1 region. The exact genomic locations of all amplicons are listed in Additional file 7. The potential interaction of endogenous GLI3 proteins with these critical amplicons was analysed by ChIP-qPCR analysis of wild-type (open bars) and Gli3Xt/Xt (black) limb buds at E11.5. Two stars indicate significant enrichment of amplicon “d” in the HMCO1 region (p = 0.008). (B) GLI3 ChIP-qPCR analysis of the HMCO2 region. Amplicons “f” and “g” are located inside HMCO2 and the previously identified GBR3 [32]. (C) GLI3 ChIP-qPCR analysis of HMCO3. All values are shown as mean ± SD.
Spatio-temporal Grem1 expression is regulated by the interaction of multiple cis-regulatory regions far downstream of the transcription unit
Previous genetic analysis showed that Grem1 transcription in limb buds is initiated by BMP signalling and up-regulated under the influence of SHH and AER-FGF signaling as limb bud development progresses [11]. Finally, Grem1 expression is terminated concomitantly with the initiation of digit formation by high levels of FGF signal transduction and GLI3-mediated repression in the anterior mouse limb bud mesenchyme [26,40]. This dynamic spatio-temporal regulation of Grem1 expression indicates that the activity of different cis-regulatory elements may change over time. The spatio-temporal activity of the GRS1 transgene (Figure 4B) indicates that it is primarily regulated by the SHH/GREM1/AER-FGF feedback signaling system as limb bud development progresses. In particular, its activity is first apparent when the SHH/GREM1/AER-FGF feedback is already established, and the expected termination does not occur, as ß-galactosidase activity remains in the posterior mesenchyme after the endogenous Grem1 transcripts have been down-regulated (Figure 4B, D). These results indicate that FGF-mediated termination of Grem1 expression and the underlying self-regulatory feedback signalling system [40] does not occur by FGF signal transduction impacting the GRS1 region. In addition, neither the 70 kb critical region of the Grem1 landscape (Grem1-LacZ, Figure 2A) nor transgenes derived from this region (Figures. 234 and ref. [32]) are able to accurately recapitulate the entire spatio-temporal distribution of Grem1 transcript in the limb bud mesenchyme. Therefore, either the interactions among these elements or additional as yet unknown cis-regulatory regions located outside the critical region must also participate in the regulation of Grem1 expression. Indeed, there is circumstantial evidence for the latter, as deletion of the GC-rich Fmn1 coding exon 9 (located ~200 kb downstream of the Grem1 transcription unit) results in non-complementation with a Grem1 null allele, causing a hypomorphic Grem1 limb skeletal phenotype, and significantly reduced Grem1 expression (ref. [29] and A.Z and R.Z., unpublished data).
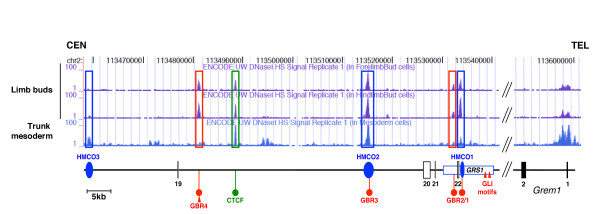
Mapping of DNAse I hypersensitive (HS) sites is used to identify active regulatory regions in chromatin (reviewed in ref. [48]). Recently, the ENCODE consortium has done genome-wide mapping of HS sites in several mouse tissues including limb buds (ENCODE Group, University of Washington) [33,34]. Interestingly, this analysis revealed three HS sites within the GBRs of the GRS1 and HMCO2, while no HS sites mapping to HMCO3 were detected in limb buds at E11.5 (Figure 8). The five HS sites mapped by the ENCODE consortium in the Grem1 genomic landscape overlap with the cis-regulatory elements required for Grem1 expression in mouse limb buds (Figure 8). The HS site mapping to HMCO1 overlaps well with the amplicon enriched by GLI3-ChIP analysis (Figure 7A). In fact, all HS sites in the Grem1 genomic landscape overlap either with the GBR regions interacting with GLI3 [26,32] or CTCF binding sites [31]. Analysis of Ctcf-deficient limb buds revealed its requirement for up-regulation and distal-anterior expansion of Grem1 expression [31]. Interestingly, the HS site overlapping with GBR4 [32] is not conserved between mammals and birds (Figure 8 and data not shown). While the early GBR4 activity is comparable to GRS1 in the posterior mesenchyme (Figure 4B), it does not support distal-anterior expansion of the expression during subsequent development [32]. Thus, the interaction of the GBR4 and GRS1 cis-regulatory regions could provide Grem1 expression with the necessary robustness and evolutionary plasticity, as has been postulated for the transcriptional regulation of HoxD genes by multiple interacting regulatory regions during limb bud development [8]. Taken together, these studies establish that the five HS sites in the Grem1 genomic landscape are functionally relevant for Grem1 expression in mouse limb buds, but not all of them are evolutionary highly conserved. Indeed, experimental and comparative evolutionary evidence indicates that alterations in the spatio-temporal expression of Grem1, and thereby the activity of the SHH/GREM1/AER-FGF feedback loop, likely contributes significantly to variations in digit numbers and morphologies (reviewed in ref. [49]).
Figure 8.
The Grem1 genomic landscape harbours multiple distant cis-regulatory regions that orchestrate Grem1 transcription in limb buds. Mapping of the DNAse I hypersensitive (HS) sites in fore-, hindlimb bud and trunk mesoderm of mouse embryos at E11.5 (ENCODE Group, University of Washington, [33,34]). A schematic representation of the cis-regulatory landscape that regulates Grem1 expression in the limb bud mesenchyme is shown below the HS site tracks. Interestingly, the major HS sites co-localise with regions that are functionally required, highly conserved (HMCO1-3, blue), and/or overlap with experimentally defined GBRs (red, ref. [32] and this study) or a CTCF binding region (green, [31]). Red arrowheads indicate consensus GLI binding sites identified by in silico analysis. HMCO1 and HMCO2 overlap with limb bud specific HS sites, while HMCO3 overlaps with a trunk mesodermal HS site. For exact coordinates, see Additional file 8.
Conclusions
In this study, we identify GRS1 as a distant cis-regulatory region that encompasses the highly conserved HMCO1 region and show that it controls important aspects of Grem1 expression in the posterior and distal-anterior limb bud mesenchyme. GRS1 activity depends critically on SHH signalling and endogenous GLI3 protein complexes interact with specific regions within HMCO1 in wild-type limb buds. In addition, the anterior expansion of GRS1 activity is also regulated by BMP signal transduction. These results together with previous studies reveal the large genomic architecture composed of several distant cis-regulatory regions that control the highly dynamic Grem1 expression in response to signalling inputs from the BMP, SHH and FGF pathways. It is likely that the interactions among these dispersed cis-regulatory regions provide the dynamic Grem1 expression in limb buds with the necessary robustness, but at the same time allow for evolutionary plasticity of its expression [8].
Methods
Ethics Statement
All the experiments were conducted in strict accordance with Swiss law following the 3R principles and the conduct defined by the Basel Declaration. All studies involving mice were classified as grade zero by the Animal Welfare and Ethics Commission of both cantons of Basel and Argovia, which implies minimal suffering.
Mouse strains
The Shh loss-of-function allele [50], the Gli3 extra-toes-J allele [51], the Grem1 null allele [13] and the GRS1tg/+ alleles (this study) were maintained in an NMRI background, while the Smad4 conditional allele (Smad4flox, [45]) and the Prx1-Cre transgene [46] were maintained in a C57BL/6 J background.
BAC modifications
BAC constructs were engineered using ET cloning as previously described [7]. The deletions ΔHMCO1, ΔHMCO2 and ΔHMCO3 were induced in the Grem1-LacZ BAC clone using a Kanamycin resistance selection cassette. All primer sequences are available upon request. See Additional file 8 for the genomic coordinates of HMCO1-3.
ßglob-LacZ transgene constructs
An expression cassette was constructed in pKS-Bluescript using LacZeocin reporter under control of the minimal human ß-globin promoter sequences [52]. Various combinations of HMCO sequences were inserted downstream into this cassette. The HMCO1, -2 and -3 core sequences were amplified by PCR directly from mouse genomic DNA (primers available on request). The 9 kb GRS1 region was initially subcloned as a 9.6 kb Bgl2 fragment from BAC RP23-113 H17 (chr2: 113,611,499-113,847,299 (mm10). BacPac Resources, Children’s Hospital, Oakland, USA). All constructs were linearized with Ksp1 before microinjection.
Generation of transgenic founder embryos and transgenic strains
BAC and ßglob-LacZ transgenic constructs were injected into the pronucleus of fertilized mouse eggs. Several founder embryos for each construct were scored for ß-galactosidase activity (Additional file 3). For the GRS1ßglob-LacZ transgene, seven independent transgenic mouse strains were established by crossing founders into the NMRI background. Transgene copy numbers were determined by real time qPCR [53] using the Bio-Rad CFX96 Real-Time PCR System in combination with the iQ SYBR Green Supermix (Bio-Rad). For each mouse, 20 ng of genomic DNA were analysed in triplicate. The primers to amplify the DBH genomic region were used for normalization and non-transgenic littermate DNA was used as control. The normalized control levels were set to 2 as the primers also amplified both alleles of the endogenous HMCO1 regions. The 2-ΔΔCt formula was used to calculate and normalize transgene copy numbers. In addition to three different F1 males, three embryos from the F2 and three embryos from the F3 generation were analysed for the two transgenic strains shown to ascertain stable transmission of the transgenes. Primers sequences are available on request.
Detection of ß-galactosidase activity
Embryos were isolated in ice-cold PBS and staged according to somites numbers, then fixed in 1% formaldehyde, 0.2% glutaraldehyde, 0.02% NP40, 0.01% sodium deoxycholate in 1x PBS for 20–30 minutes at 4°C. Subsequently, embryos were washed three times in 1x PBS for 5 minutes at room temperature and incubated overnight at 37°C and in the dark in 1 mg/mL X-Gal, 0.25 mM K3Fe(CN6), 0.25 mM K4Fe(CN6), 0.01% NP-40, 0.4 mM MgCl2 and 1% sodium deoxycholate to detect ß-galactosidase activity, which colours cells blue. To stop the reaction, embryos were washed three times in 1x PBS for 5 minutes each at room temperature.
Chromatin Immunoprecipitation (ChIP)
Forelimbs and hindlimbs of 10 wild-type or Gli3Xt/Xt embryos at E11.5 were dissected and processed for ChIP as described [26] using a polyclonal anti-GLI3 antibody (#2676, Genentech, [47]). To compute the level of enrichment of a given region, the Ct values of both input and ChIP samples were compared with those of a negative control amplicon located in the mouse ß-actin locus [21]. All results (mean ± SD) were obtained by analysing three completely independent experiments per genotype. The significance of all differences was assessed using the two-tailed, non-parametric Mann–Whitney test. The coordinates of the relevant amplification are shown in Additional file 7.
All genomic sequence alignements of the Fmn1-Grem1 locus region were performed using the ECR browser [54] and the ClustalW2 program [55].
Authors’ contributions
AZ conceived most of the experiments and wrote the manuscript together with RZ. AZ also carried out most of the genetic and transgenic analysis together with FL. FL also prepared the figures for the manuscript. FL, JLR and NM performed the in silico analysis. JLR carried out the ChIP analysis to detect the endogenous GLI3 protein complexes. NM contributed in the initial phase of this project (generation of transgenic constructs and initial analysis of founder embryos). CK generated the transgenic founders for all constructs. RZ conceived part of the study and wrote the manuscript together with AZ. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Conservation of the HMCO core regions. ClustalW2 multiple sequence alignment of the HMCO1, HMCO2 and HMCO3 core regions of mouse (mm10), human (hg19), chimpanzee (panTro3), dog (canFam2), bovine (bosTau6), opossum (monDom5), chicken (galGal3), lizard (anoCar2), frog (xenTro2), and coelacanth (LatCha1) genomes The corresponding genomic coordinates are indicated. HMCO1: 73 of 149 nucleotides were conserved in all species; HMCO2: 182/298 conserved nucleotides, HMCO3: 45/137 conserved nucleotides.
Table S1. Genomic coordinates for the sequence comparisons shown in Figure 1A.
Table S2. Analysis of transgenic founder embryo.
Figure S2. Limb bud mesenchymal expression of GRS1-ßglob-LacZ transgene in two independent transgenic mouse strains. (A) Expression of the GRS1-ßglob-LacZ transgene is restricted to limb buds, with expression initiating earlier in forelimb than hindlimb buds. Note the differences in ß-galactosidase activity in GRS1tg10/+ (left panel) and GRS1tg2/+ (right panel) transgenic mouse embryos. (B) Using real-time qPCR, the transgene copy numbers in both the GRS1tg10/+ and GRS1tg2/+ mouse strains were determined in comparison to wild-type mice (carrying 2 copies of the endogenous GRS1 regions). This analysis revealed that the GRS1tg10/+ strain carries 10 copies and GRS1tg2/+ 2 copies of the transgene, respectively.
Figure S3. Comparison of the LacZ mRNA and ß-galactosidase reporter activity in early limb buds of GRS1tg10/+ embryos. Distribution of ß-galactosidase activity (A) and LacZ transcripts (B) in forelimb buds of GRS1tg10/+ embryos at E10.0 and E10.5. Arrowheads point to the posterior expression domains.
Figure S4. Dorso-ventral distribution of ß-galactosidase activity in forelimb buds expressing the GRS1tg2/+ transgene. The GRS1tg2/+ transgene respects the dorsal and ventral restriction of the mesenchymal expression domains. Note that overall expression is significantly lower than in GRS1tg10/+ limb buds (compare to Figure 5B). D: dorsal, V: ventral.
Table S3. qPCR amplicons for GLI3 ChIP analysis.
Table S4. Genomic coordinates of identified Grem1 regulatory regions.
Contributor Information
Aimée Zuniga, Email: aimee.zuniga@unibas.ch.
Frédéric Laurent, Email: frederic.laurent@unibas.ch.
Javier Lopez-Rios, Email: Javier.Lopez-Rios@unibas.ch.
Christian Klasen, Email: Christian.Klasen@mdc-berlin.de.
Nicolas Matt, Email: n.matt@ibmc.u-strasbg.fr.
Rolf Zeller, Email: rolf.zeller@unibas.ch.
Acknowledgements
The authors are grateful to A. Offinger and her staff for excellent animal care. L. d'Amato and Christina Torres are thanked for technical assistance. S. Scales (Genentech) provided the GLI3 antibodies. We thank group members for critical input on the manuscript. The ENCODE Group at University of Washington (J. Stamatoyannopoulos and coworkers) are acknowledged for the genome-wide DNase I hypersensitive site mapping data made available freely through the ENCODE Data Coordination Center at UCSC (release date on March 29, 2012). NM received a EU Marie Curie postdoctoral fellowship. This research is supported by grants of the Swiss National Science Foundation (grants no. 31003A_130803 to RZ and 310000_122558 to AZ), a EU reintegration grant (PERG-GA-2009-246576 to JLR) and the University of Basel. The authors declare no conflict of interest.
References
- Gordon CT, Tan TY, Benko S, Fitzpatrick D, Lyonnet S. et al. Long-range regulation at the SOX9 locus in development and disease. Journal of medical genetics. 2009;46:649–656. doi: 10.1136/jmg.2009.068361. [DOI] [PubMed] [Google Scholar]
- Vandermeer JE, Ahituv N. cis-regulatory mutations are a genetic cause of human limb malformations. Dev Dyn. 2011;240:920–930. doi: 10.1002/dvdy.22535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga A, Probst S, Zeller R. The molecular basis of human congenital limb malformations. Wiley Interdisciplinary Reviews: Developmental Biology; 2012. [DOI] [PubMed] [Google Scholar]
- Ruf S, Symmons O, Uslu VV, Dolle D, Hot C. et al. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat Genet. 2011;43:379–386. doi: 10.1038/ng.790. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Horikoshi T, Heaney SJ, Van Baren MJ, Van Der Linde HC. et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci U S A. 2002;99:7548–7553. doi: 10.1073/pnas.112212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/S0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Zuniga A, Michos O, Spitz F, Haramis AP, Panman L. et al. Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression. Genes Dev. 2004;18:1553–1564. doi: 10.1101/gad.299904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L. et al. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Spitz F. Control of vertebrate Hox clusters by remote and global cis-acting regulatory sequences. Adv Exp Med Biol. 2010;689:63–78. doi: 10.1007/978-1-4419-6673-5_4. [DOI] [PubMed] [Google Scholar]
- Zeller R, Zuniga A. Shh and Gremlin1 chromosomal landscapes in development and disease. Curr Opin Genet Dev. 2007;17:428–434. doi: 10.1016/j.gde.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Benazet JD, Bischofberger M, Tiecke E, Goncalves A, Martin JF. et al. A self-regulatory system of interlinked signaling feedback loops controls mouse limb patterning. Science. 2009;323:1050–1053. doi: 10.1126/science.1168755. [DOI] [PubMed] [Google Scholar]
- Capdevila J, Tsukui T, Rodriquez Esteban C, Zappavigna V, Izpisua Belmonte JC. Control of vertebrate limb outgrowth by the proximal factor Meis2 and distal antagonism of BMPs by Gremlin. Mol Cell. 1999;4:839–849. doi: 10.1016/S1097-2765(00)80393-7. [DOI] [PubMed] [Google Scholar]
- Michos O, Panman L, Vintersten K, Beier K, Zeller R. et al. Gremlin-mediated BMP antagonism induces the epithelial-mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development. 2004;131:3401–3410. doi: 10.1242/dev.01251. [DOI] [PubMed] [Google Scholar]
- Zuniga A, Haramis AP, McMahon AP, Zeller R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 1999;401:598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]
- Zeller R, Lopez-Rios J, Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet. 2009;10:845–858. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
- Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Hill AE, Devenney PS, Hill RE. Point mutations in a distant sonic hedgehog cis-regulator generate a variable regulatory output responsible for preaxial polydactyly. Hum Mol Genet. 2008;17:978–985. doi: 10.1093/hmg/ddm370. [DOI] [PubMed] [Google Scholar]
- Maas SA, Suzuki T, Fallon JF. Identification of spontaneous mutations within the long-range limb-specific Sonic hedgehog enhancer (ZRS) that alter Sonic hedgehog expression in the chicken limb mutants oligozeugodactyly and silkie breed. Dev Dyn. 2011;240:1212–1222. doi: 10.1002/dvdy.22634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H. et al. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell. 2009;16:47–57. doi: 10.1016/j.devcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Capellini TD, Di Giacomo G, Salsi V, Brendolan A, Ferretti E. et al. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development. 2006;133:2263–2273. doi: 10.1242/dev.02395. [DOI] [PubMed] [Google Scholar]
- Galli A, Robay D, Osterwalder M, Bao X, Benazet JD. et al. Distinct roles of Hand2 in initiating polarity and posterior Shh expression during the onset of mouse limb bud development. PLoS Genet. 2010;6:e1000901. doi: 10.1371/journal.pgen.1000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Williamson I, Wiltshire JH, Peluso S, Devenney PS. et al. Opposing Functions of the ETS Factor Family Define Shh Spatial Expression in Limb Buds and Underlie Polydactyly. Dev Cell. 2012;22:459–467. doi: 10.1016/j.devcel.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-Regulated Processing of Gli3 Produces an Anterior/Posterior Repressor gradient in the Developing Vertebrate Limb. Cell. 2000;100:423–434. doi: 10.1016/S0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Mo R, Freer AM, Zinyk DL, Crackower MA, Michaud J. et al. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124:113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- Litingtung Y, Dahn RD, Li Y, Fallon JF, Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418:979–983. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- Lopez-Rios J, Speziale D, Robay D, Scotti M, Osterwalder M. et al. GLI3 Constrains Digit Number by Controlling Both Progenitor Proliferation and BMP-Dependent Exit to Chondrogenesis. Developmental Cell. 2012;22:837–848. doi: 10.1016/j.devcel.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Welscher P, Zuniga A, Kuijper S, Drenth T, Goedemans HJ. et al. Progression of Vertebrate Limb Development through SHH-Mediated Counteraction of GLI3. Science. 2002;298:827–830. doi: 10.1126/science.1075620. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Hsu D, Brunet LJ, Dionne MS, Harland RM. Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat Genet. 2003;34:303–307. doi: 10.1038/ng1178. [DOI] [PubMed] [Google Scholar]
- Zhou F, Leder P, Zuniga A, Dettenhofer M. Formin1 disruption confers oligodactylism and alters Bmp signaling. Hum Mol Genet. 2009;18:2472–2482. doi: 10.1093/hmg/ddp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panman L, Galli A, Lagarde N, Michos O, Soete G. et al. Differential regulation of gene expression in the digit forming area of the mouse limb bud by SHH and gremlin 1/FGF-mediated epithelial-mesenchymal signalling. Development. 2006;133:3419–3428. doi: 10.1242/dev.02529. [DOI] [PubMed] [Google Scholar]
- Soshnikova N, Montavon T, Leleu M, Galjart N, Duboule D. Functional analysis of CTCF during mammalian limb development. Dev Cell. 2010;19:819–830. doi: 10.1016/j.devcel.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Vokes SA, Ji H, Wong WH, McMahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22:2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC. et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS biology. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl EJ, Barker D, Day RC, Beck CW. Identification of genes associated with regenerative success of Xenopus laevis hindlimbs. BMC Dev Biol. 2008;8:66. doi: 10.1186/1471-213X-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoli S, Gilardelli CN, Pozzoli O, Presta M, Cotelli F. Regulated expression pattern of gremlin during zebrafish development. Gene expression patterns : GEP. 2005;5:539–544. doi: 10.1016/j.modgep.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Boisvert CA, Mark-Kurik E, Ahlberg PE. The pectoral fin of Panderichthys and the origin of digits. Nature. 2008;456:636–638. doi: 10.1038/nature07339. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P. et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Scherz PJ, Harfe BD, McMahon AP, Tabin CJ. The limb bud Shh-Fgf feedback loop is terminated by expansion of former ZPA cells. Science. 2004;305:396–399. doi: 10.1126/science.1096966. [DOI] [PubMed] [Google Scholar]
- Verheyden JM, Sun X. An Fgf/Gremlin inhibitory feedback loop triggers termination of limb bud outgrowth. Nature. 2008;454:638–641. doi: 10.1038/nature07085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, Tsuji K, Cox K, Harfe BD, Rosen V. et al. Genetic Analysis of the Roles of BMP2, BMP4, and BMP7 in Limb Patterning and Skeletogenesis. PLoS Genet. 2006;2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzchori I, Day TF, Carolan PJ, Zhao Y, Wassif CA. et al. LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development. 2009;136:1375–1385. doi: 10.1242/dev.026476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim S, Hasso SM, Fallon JF, Tabin CJ. Regulation of Gremlin expression in the posterior limb bud. Dev Biol. 2006;299:12–21. doi: 10.1016/j.ydbio.2006.05.026. [DOI] [PubMed] [Google Scholar]
- te Welscher P, Fernandez-Teran M, Ros MA, Zeller R. Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prior to SHH signaling. Genes Dev. 2002;16:421–426. doi: 10.1101/gad.219202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li C, Herrera PL, Deng CX. Generation of Smad4/Dpc4 conditional knockout mice. Genesis. 2002;32:80–81. doi: 10.1002/gene.10029. [DOI] [PubMed] [Google Scholar]
- Logan M, Martin JF, Nagy A, Lobe C, Olson EN. et al. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ. et al. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Molecular and cellular biology. 2010;30:1910–1922. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nature reviews Genetics. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller R. The temporal dynamics of vertebrate limb development, teratogenesis and evolution. Curr Opin Genet Dev. 2010;20:384–390. doi: 10.1016/j.gde.2010.04.014. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Dassule HR, Karavanova I, Botchkarev VA, Li J. et al. Sonic hedgehog signaling is essential for hair development. Curr Biol. 1998;8:1058–1068. doi: 10.1016/S0960-9822(98)70443-9. [DOI] [PubMed] [Google Scholar]
- Maynard TM, Jain MD, Balmer CW, LaMantia AS. High-resolution mapping of the Gli3 mutation extra-toes reveals a 51.5-kb deletion. Mamm Genome. 2002;13:58–61. doi: 10.1007/s00335-001-2115-X. [DOI] [PubMed] [Google Scholar]
- Morgan BA, Conlon FL, Manzanares M, Millar JB, Kanuga N. et al. Transposon tools for recombinant DNA manipulation: characterization of transcriptional regulators from yeast, Xenopus, and mouse. Proc Natl Acad Sci U S A. 1996;93:2801–2806. doi: 10.1073/pnas.93.7.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson L, Heslan JM, Menoret S, Anegon I. Rapid and accurate determination of zygosity in transgenic animals by real-time quantitative PCR. Transgenic research. 2002;11:43–48. doi: 10.1023/A:1013928600442. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic acids research. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Conservation of the HMCO core regions. ClustalW2 multiple sequence alignment of the HMCO1, HMCO2 and HMCO3 core regions of mouse (mm10), human (hg19), chimpanzee (panTro3), dog (canFam2), bovine (bosTau6), opossum (monDom5), chicken (galGal3), lizard (anoCar2), frog (xenTro2), and coelacanth (LatCha1) genomes The corresponding genomic coordinates are indicated. HMCO1: 73 of 149 nucleotides were conserved in all species; HMCO2: 182/298 conserved nucleotides, HMCO3: 45/137 conserved nucleotides.
Table S1. Genomic coordinates for the sequence comparisons shown in Figure 1A.
Table S2. Analysis of transgenic founder embryo.
Figure S2. Limb bud mesenchymal expression of GRS1-ßglob-LacZ transgene in two independent transgenic mouse strains. (A) Expression of the GRS1-ßglob-LacZ transgene is restricted to limb buds, with expression initiating earlier in forelimb than hindlimb buds. Note the differences in ß-galactosidase activity in GRS1tg10/+ (left panel) and GRS1tg2/+ (right panel) transgenic mouse embryos. (B) Using real-time qPCR, the transgene copy numbers in both the GRS1tg10/+ and GRS1tg2/+ mouse strains were determined in comparison to wild-type mice (carrying 2 copies of the endogenous GRS1 regions). This analysis revealed that the GRS1tg10/+ strain carries 10 copies and GRS1tg2/+ 2 copies of the transgene, respectively.
Figure S3. Comparison of the LacZ mRNA and ß-galactosidase reporter activity in early limb buds of GRS1tg10/+ embryos. Distribution of ß-galactosidase activity (A) and LacZ transcripts (B) in forelimb buds of GRS1tg10/+ embryos at E10.0 and E10.5. Arrowheads point to the posterior expression domains.
Figure S4. Dorso-ventral distribution of ß-galactosidase activity in forelimb buds expressing the GRS1tg2/+ transgene. The GRS1tg2/+ transgene respects the dorsal and ventral restriction of the mesenchymal expression domains. Note that overall expression is significantly lower than in GRS1tg10/+ limb buds (compare to Figure 5B). D: dorsal, V: ventral.
Table S3. qPCR amplicons for GLI3 ChIP analysis.
Table S4. Genomic coordinates of identified Grem1 regulatory regions.