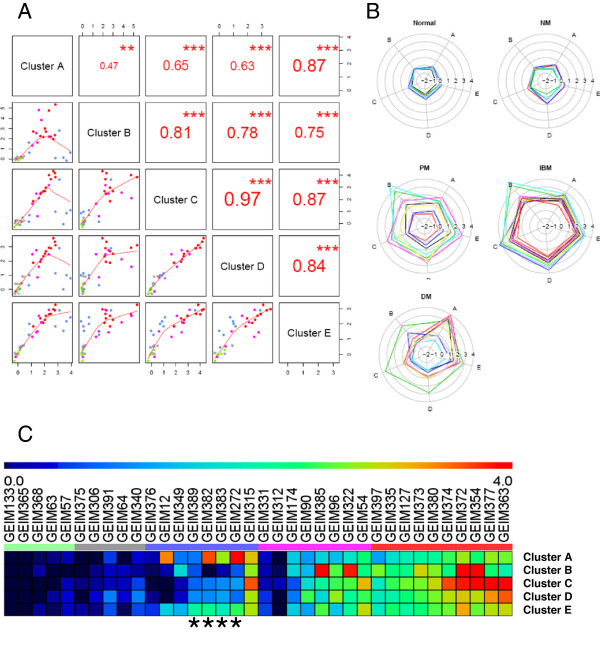
Figure 2.
Gene signatures derived from clusters A-E from the consensus cluster. (A) Pairwise scatter plots of the five gene signatures. The scatter plots of the signatures of the clusters A-E are displayed in the lower panels, where data points are colored by the sample type (same as the Figure 1) and red lines represent the lowess (Locally Weighted Scatterplot Smoothing) lines. The corresponding spearman correlation coefficients are displayed in the upper panels, with significance levels indicated by the stars (*** p < 0.001, ** p < 0.01, * p < 0.05). (B) Subclasses of myositis characterized by the five gene signatures are shown in the star plots. Each subject is represented with a specific pentagon in the star plot and the vertices of one pentagon along the spokes (A-E) represent enrichment of the signature scores from clusters A-E. Colors of pentagons are randomly assigned to distinguish subjects and star plots are grouped by the myositis subclasses. (C) Subjects can be further characterized by the five gene signatures using a heatmap. The signature scores are represented by the rainbow color in the heatmap, ranging between 0 (colored in blue) and 4 (colored in red); the color key for each sample’s disease type is displayed horizontally between the sample identifiers and the heatmap, where, (from left to right), green denotes normal, gray for NM, blue for DM, magenta for PM and red for IBM. The four DM patients with perifascicular atrophy (PFA) are marked by the star symbol.