Abstract
Technological innovations and translation of basic discoveries to clinical practice drive advances in medicine. Today's innovative technologies enable comprehensive screening of the genome, transcriptome, proteome, and metabolome. The detailed knowledge, converged in the integrated “omics” (genomics, transcriptomics, proteomics, and metabolomics), holds an immense potential for understanding mechanism of diseases, facilitating their early diagnostics, selecting personalized therapeutic strategies, and assessing their effectiveness. Metabolomics is the newest “omics” approach aimed to analyze large metabolite pools. The next generation of metabolomic screening requires technologies for high throughput and robust monitoring of metabolite levels and their fluxes. In this regard, stable isotope 18O-based metabolite tagging technology expands quantitative measurements of metabolite levels and turnover rates to all metabolites that include water as a reactant, most notably phosphometabolites. The obtained profiles and turnover rates are sensitive indicators of energy and metabolic imbalances like the ones created by genetic deficiencies, myocardial ischemia, heart failure, neurodegenerative disorders, etc. Here we describe and discuss briefly the potential use of dynamic phosphometabolomic platform for disease diagnostics currently under development at Mayo Clinic.
Integrated “omics” approach
Living cells represent an integrated and interacting network of genes, transcripts, proteins, small signaling molecules, and metabolites that define cellular phenotype and function. Traditionally the focus of biomedical research was on individual genes, single protein targets, single metabolites, and metabolic or signaling pathways. This “molecular reductionist” paradigm was based on the assumption that identifying genetic variations and molecular components would lead to discovery of cures for human diseases. However, most of diseases are complex and multi-factorial and the disease phenotype is determined by the alterations of multiple genes, pathways, proteins and metabolites (at cellular, tissue, and organismal levels). Therefore, an integrated “omics” approach is more viable direction for uncovering alterations in metabolic networks, disease mechanisms, and mechanisms of drug effects.
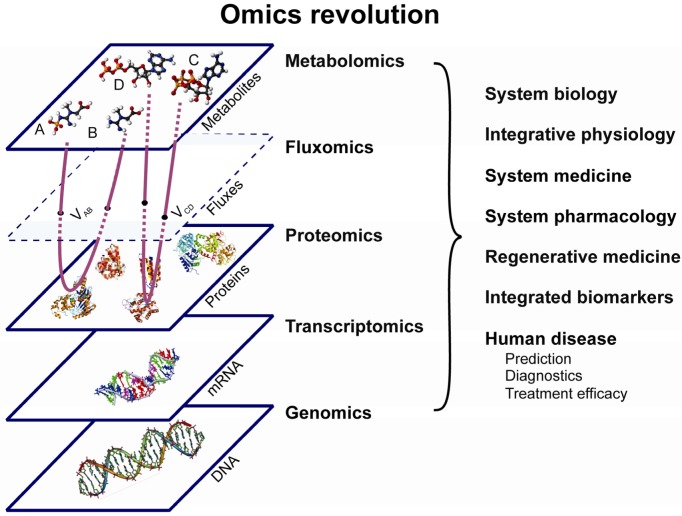
Recent advent of large-scale metabolomics and fluxomic (metabolite dynamics and metabolic flux analysis) completed the “omics revolution” (Figure 1), where genomics, transcriptomics, proteomics, metabolomics, and fluxomics all together complement phenotype determination of living organism. Such integrated “omics” cascades provide a framework for advances in system and network biology, integrative physiology, and system medicine as well as system pharmacology and regenerative medicine. Noteworthy is the “reverse omic” approach or “metabolomics-informed pharmacogenomics,” where discovery of specific metabolite changes have led to discovery of genetic alterations (2). Therefore, bringing new “omics” technologies to clinical practice will improve disease diagnostics and treatment by targeting drugs and procedures for each unique transcriptomic and metabolomic profiles.
Figure 1.
The “omics revolution” – an integrated comprehensive “omics” approach combining genomics, transcriptomics, proteomics, metabolomics, and fluxomics for advancement of systemic sciences and for human disease diagnostics and treatment. After Nielsen and Oliver (1).
Dynamic metabolomic technologies
Comprehensive characterization of metabolic networks and their function requires quantitative knowledge of metabolite concentrations (metabolomics) and metabolite fluxes (fluxomics). Analytically, this could be perceived as a determination of concentrations and turnover rates of a large number of small molecules (metabolites) from tissue or body fluids. Thus, metabolomics and fluxomics methods heavily rely on the information-rich analytical techniques, most notably nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry. Stable isotope 18O-assisted 31P NMR and mass spectrometry provide means for simultaneous measurements of phosphorous-containing metabolite (phosphometabolite) levels and their respective turnover rates in tissue and blood samples. The 18O labeling of metabolites is based on the incorporation of the 18O nuclei (from H218O), into phosphate group with each act of adenosine triphosphate (ATP) hydrolysis, and subsequent distribution of 18O-labeled phosphoryls among phosphate-carrying molecules. 18O is a natural and stable isotope of oxygen. When tissue or cells are exposed to media containing water with a known percentage of 18O, H218O rapidly equilibrates with the cellular water, and 18O from water is incorporated into cellular phosphate metabolites proportionally to the rate of the involved enzymatic reactions (3). All major phosphometabolites and their turnover rates can be quantified using 18O-assisted 31P NMR spectroscopy and much faster by the use of mass spectrometry. The 18O isotope effect is readily visible as 18O induced 31P chemical shift in high-resolution 31P NMR spectra, and as 2 amu change of mass in mass spectra. Thus, stable isotope tracer-based metabolomic technologies allow simultaneous quantitative determination of metabolite levels and their turnover rates with subsequent evaluation of metabolic network dynamics.
Dynamic phosphmetabolomic platform under development at Mayo Clinic (Rochester, MN, USA) is shown in Table 1. This analytical platform includes metabolome determination, providing static metabolic profile, and dynamic phosphometabolomics where phosphometabolite turnover rates, metabolic flux distribution in phosphotransfer networks, and dynamics of metabolic cycles are analyzed. Based on the data obtained by these technologies, a correlation matrix and predictive algorithm is being developed for disease diagnostics.
Table 1.
Dynamic phosphmetabolomic platform*
| Metabolomics → | Dynamic phosphometabolomics (stable isotope 18O-labeling) → | Clinical metabolomics and 18O phosphometabolomics |
|---|---|---|
| Metabolome, static metabolic profile. Technologies: Tissue, cell and body fluid samples GC/MS LC/MS 1H NMR 31P NMR | Phosphometabolite turnover rates, metabolic flux distribution analysis in phosphotransfer networks and dynamics of metabolic cycles. Technologies: 18O-labeling of tissue, cell and whole blood samples 18O-assisted GC/MS 18O-assisted LC/MS 18O-assisted 31P NMR Model-based flux analysis | Metabolomics + Dynamic phosphometabolomics = system and network approach, disease mechanisms, more accurate disease prediction, diagnosis, treatment choices and efficacy. Technologies: 18O-labeling of tissue, cell and whole blood samples 18O-assisted GC/MS 18O-assisted LC/MS Correlation and predictive algorithms |
*Abbreviations: GC – gas chromatography; MS – mass spectrometry; LC – liquid chromatography; NMR – nuclear magnetic resonance.
Dynamic metabolomic profiling
The analysis of metabolic fingerprints left by disease processes and metabolic monitoring of disease progression or treatment efficacy plays a crucial role in personalized and predictive medicine. Metabolomic profiling may not be enough to predict the phenotype as it gives only an instant static picture of the physiology of a living organism. To entirely understand metabolic phenotypes and network dynamics, quantitative knowledge of metabolite turnover rates is required (4-13). This is because significant alterations in metabolic flux could take place without obvious changes in metabolite concentrations, especially metabolites associated with high flux/turnover rates (6,14). Stable isotope tracer-based metabolomic technologies allow for simultaneous determination of metabolite levels and their turnover rates with subsequent evaluation of metabolic network dynamics (10,11,13-16). For example, 13C labeling is widely used to track turnover of the carbon backbone of metabolites and label propagation through metabolic networks (17-19). However, it does not provide data on the status of the phosphate-containing metabolite based cellular energetic. On the other hand, 18O isotopes are conveniently used to follow phosphate transfer rates of energetically and signal transduction important biomolecules (3,10,16,20-24). As water is involved in many enzymatic reactions, 18O from the H218O can be incorporated into array of metabolites. This expands the use of 18O technology to labeling of many non-phosphate containing metabolites, which in turn enables observing of large metabolic networks in intact tissues and alterations in human disease. In this regard, 18O stable isotope labeling is widely used in quantitative proteomics to detect changes in protein levels and in their turnover rates (25,26). We believe that the analysis of phosphometabolite turnover rates using 18O labeling (in whole fresh blood, blood cell fractions, plasma, and tissue samples) will soon become practical enough to be routinely used in translational research, metabolic phenotyping, biomarker development, and ultimately in diagnostics and treatment of human diseases.
Dynamic phosphometabolomics
Phosphate is indispensable to life activity and is the most common fragment in terms of the frequency of occurrence in the metabolome of living organisms (27,28). Phosphometabolite dynamics can be a predictor of atrial fibrillation and prothrombotic events critical in prevention of stroke and myocardial infarction (29). 18O-assisted gas chromatography/mass spectrometry (GC/MS), liquid chromatography/mass spectrometry (LC/MS), and 31P NMR technologies fill a critical gap in metabolomics technologies by providing a currently unavailable tool for analysis of perturbations in cellular energetic and metabolic signaling networks induced by diseases or genetic and acquired metabolic deficiencies (29). It can be further developed to clinically-useful stable-isotope based bioanalytical platform for analysis of phosphometabolite levels and turnover rates with high precision and extended capacity for molecular recognition and automated isotopomer analysis (30,31).
Phosphometabolomics is a new emerging area in metabolomic analyses targeting over 400 phosphometabolites critical in energetic and signaling processes. Currently, no single analytical tool fulfills all requirements for an ideal phosphometabolomic profiling, due to the physicochemical diversity of phosphometabolites, from hydrophobic phospholipids to hydrophilic phosphocarbohydrates, and phosphoamino- and non-phosphoamino-organic acids. Thus, different analytical techniques must be used to generate a comprehensive metabolomics profile (7,32,33). We established a dynamic phosphometabolomic platform that includes 18O-assisted GC/MS, 18O-assisted 31P NMR, together with 1H NMR and high pressure liquid chromatography (HPLC) (29). 18O-assisted GC/MS technology allows separation and quantitation of 18O/16O isotope ratios in phosphoryl metabolites with a molecular mass <500 Da. 18O-labeling of higher molecular weight phosphates and oligophosphates, such as ATP, can be analyzed by LC/MS and 31P NMR. 2D 1H-31P NMR total correlation spectroscopy (TOCSY) and 1D 1H NMR and HPLC complement the analysis, identification, and fractionation of phosphometabolites.
The understanding of the dynamics of oligophosphates at different phosphoryl moieties provides unique knowledge of energetic, signal transduction, and biosynthetic processes in the cell (3,16,20-24,34). For instance, all 18O isotopologues in ATP can be directly translated into activity of adenylate kinase, ATP synthase, ATPase, pyrophosphokinase, nucleotidyl transferases, and other energetic enzymes (16,31,34-37). We start to develop software for automatic analysis of isotope distribution which will enable monitoring of phosphometabolite turnover rates in whole fresh blood and tissue samples in minutes. Blood plasma and cells carry a wealth of metabolic information about health status and disease biomarkers. Blood phosphometabolite and nucleotide dynamics are important in regulation of blood flow and coagulation and their alterations may precipitate the risk of thrombosis and hypertension.
Stable isotope 18O labeling is a suitable technique to follow cellular phosphate group dynamics through phosphotransfer network. Thus, the 18O-based metabolite tagging technology bridges traditional static metabolomics with fluxomics allowing dynamic analysis of metabolic networks. For example, by using 18O-assisted analytical platforms, we were able to determine dynamic metabolic signatures of pacing-induced heart failure and metabolic phenotypes of transgenic mouse models of human diseases, associated with K-ATP channels and phosphotransfer enzyme deficiencies, as well as the effects of environmental stress (such as hypoxia) on global changes in energetics and metabolic signaling networks (9,15,38-43).
A large scale 18O stable isotope-based metabolomic technology enables determination of dynamic metabolomic signatures critical in understanding mechanisms of metabolic deficiencies and for early diagnosis and monitoring of human diseases. An advanced large scale clinically-usable phosphometabolomic technology for accurate monitoring of phosphometabolite turnover rates in human tissue, plasma and whole fresh blood samples would improve disease risk assessment, diagnosis, prognosis, and treatment of body energy and metabolic imbalance diseases. Collectively, a novel stable isotope methodology (18O-based mass spectrometry and 31P NMR spectroscopy) will bring to clinical practice advanced stable isotope-based metabolomic profiling for risk stratification, prediction of disease course, and personalized treatment of diseases by targeting drugs and procedures for each individual metabolomic profile.
Acknowledgments
Funding Supported by National Institutes of Health, Marriott Heart Disease Research Program, Marriott Foundation and Mayo Clinic.
Ethical approval Not required.
Declaration of authorship EN contributed to the method development and writing of the manuscript. SZ contributed to the method development and experiments. NOJ contributed to the method development and writing of the manuscript. AT contributed to the development of concept and writing of the manuscript. SM contributed to the method development and writing of the manuscript. PD contributed to concept development, method development, and wrote the manuscript.
Competing interests All authors have completed the Unified Competing Interest form at www.icmje.org/coi_disclosure.pdf (available on request from the corresponding author) and declare: no support from any organization for the submitted work; no financial relationships with any organizations that might have an interest in the submitted work in the previous 3 years; no other relationships or activities that could appear to have influenced the submitted work.
References
- 1.Nielsen J, Oliver S. The next wave in metabolome analysis. Trends Biotechnol. 2005;23:544–6. doi: 10.1016/j.tibtech.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Ji Y, Hebbring S, Zhu H, Jenkins GD, Biernacka J, Snyder K, et al. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther. 2011;89:97–104. doi: 10.1038/clpt.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawis SM, Walseth TF, Deeg MA, Heyman RA, Graeff RM, Goldberg ND. Adenosine triphosphate utilization rates and metabolic pool sizes in intact cells measured by transfer of 18O from water. Biophys J. 1989;55:79–99. doi: 10.1016/S0006-3495(89)82782-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ratcliffe RG, Shachar-Hill Y. Revealing metabolic phenotypes in plants: inputs from NMR analysis. Biol Rev Camb Philos Soc. 2005;80:27–43. doi: 10.1017/S1464793104006530. [DOI] [PubMed] [Google Scholar]
- 5.Cornish-Bowden A, Cardenas ML. From genome to cellular phenotype – a role for metabolic flux analysis? Nat Biotechnol. 2000;18:267–8. doi: 10.1038/73696. [DOI] [PubMed] [Google Scholar]
- 6.Kruger NJ, Ratcliffe RG. Insights into plant metabolic networks from steady-state metabolic flux analysis. Biochimie. 2009;91:697–702. doi: 10.1016/j.biochi.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Weckwerth W. Metabolomics: methods and protocols: Humana Pr Inc; 2007.
- 8.Janssen E, Dzeja PP, Oerlemans F, Simonetti AW, Heerschap A, De Haan A, et al. Adenylate kinase 1 gene deletion disrupts muscle energetic economy despite metabolic rearrangement. EMBO J. 2000;19:6371–81. doi: 10.1093/emboj/19.23.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dzeja PP, Terzic A, Wieringa B. Phosphotransfer dynamics in skeletal muscle from creatine kinase gene-deleted mice. Mol Cell Biochem. 2004;256-257:13–27. doi: 10.1023/B:MCBI.0000009856.23646.38. [DOI] [PubMed] [Google Scholar]
- 10.Pucar D, Dzeja PP, Bast P, Juranic N, Macura S, Terzic A. Cellular energetics in the preconditioned state: protective role for phosphotransfer reactions captured by 18O-assisted 31P NMR. J Biol Chem. 2001;276:44812–9. doi: 10.1074/jbc.M104425200. [DOI] [PubMed] [Google Scholar]
- 11.Griffin JL, Des Rosiers C. Applications of metabolomics and proteomics to the mdx mouse model of Duchenne muscular dystrophy: lessons from downstream of the transcriptome. Genome Med. 2009;1:32. doi: 10.1186/gm32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelleher JK. Flux estimation using isotopic tracers: common ground for metabolic physiology and metabolic engineering. Metab Eng. 2001;3:100–10. doi: 10.1006/mben.2001.0185. [DOI] [PubMed] [Google Scholar]
- 13.Dzeja PP, Hoyer K, Tian R, Zhang S, Nemutlu E, Spindler M, et al. Rearrangement of energetic and substrate utilization networks compensate for chronic myocardial creatine kinase deficiency. J Physiol. 2011;589:5193–211. doi: 10.1113/jphysiol.2011.212829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul Lee WN, Wahjudi PN, Xu J, Go VL. Tracer-based metabolomics: concepts and practices. Clin Biochem. 2010;43:1269–77. doi: 10.1016/j.clinbiochem.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dzeja PP, Terzic A. Phosphotransfer networks and cellular energetics. J Exp Biol. 2003;206:2039–47. doi: 10.1242/jeb.00426. [DOI] [PubMed] [Google Scholar]
- 16.Pucar D, Dzeja PP, Bast P, Gumina RJ, Drahl C, Lim L. Mapping hypoxia-induced bioenergetic rearrangements and metabolic signaling by O-18-assisted P-31 NMR and H-1 NMR spectroscopy. Mol Cell Biochem. 2004;256-257:281–9. doi: 10.1023/B:MCBI.0000009875.30308.7a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer E, Zamboni N, Sauer U. High-throughput metabolic flux analysis based on gas chromatography-mass spectrometry derived C-13 constraints. Anal Biochem. 2004;325:308–16. doi: 10.1016/j.ab.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 18.Noh K, Gronke K, Luo B, Takors R, Oldiges M, Wiechert W. Metabolic flux analysis at ultra short time scale: Isotopically non-stationary C-13 labeling experiments. J Biotechnol. 2007;129:249–67. doi: 10.1016/j.jbiotec.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 19.Zamboni N. (13)C metabolic flux analysis in complex systems. Curr Opin Biotechnol. 2011;22:103–8. doi: 10.1016/j.copbio.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Stempel KE, Boyer PD. Refinement in oxygen-18 methodology for the study of phosphorylation mechanisms. Methods Enzymol. 1986;126:618–39. doi: 10.1016/S0076-6879(86)26065-6. [DOI] [PubMed] [Google Scholar]
- 21.Zeleznikar RJ, Goldberg ND. Kinetics and compartmentation of energy metabolism in intact skeletal muscle determined from 18O labeling of metabolite phosphoryls. J Biol Chem. 1991;266:15110–9. [PubMed] [Google Scholar]
- 22.Pucar D, Janssen E, Dzeja PP, Juranic N, Macura S, Wieringa B, et al. Compromised energetics in the adenylate kinase AK1 gene knockout heart under metabolic stress. J Biol Chem. 2000;275:41424–9. doi: 10.1074/jbc.M007903200. [DOI] [PubMed] [Google Scholar]
- 23.Pucar D, Bast P, Gumina RJ, Lim L, Drahl C, Juranic N, et al. Adenylate kinase AK1 knockout heart: energetics and functional performance under ischemia-reperfusion. Am J Physiol Heart Circ Physiol. 2002;283:H776–82. doi: 10.1152/ajpheart.00116.2002. [DOI] [PubMed] [Google Scholar]
- 24.Cohn M, Hu A. Isotopic (18O) shift in 31P nuclear magnetic resonance applied to a study of enzyme-catalyzed phosphate–phosphate exchange and phosphate (oxygen) – water exchange reactions. Proc Natl Acad Sci U S A. 1978;75:200–3. doi: 10.1073/pnas.75.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye X, Luke B, Andresson T, Blonder J. 18O stable isotope labeling in MS-based proteomics. Brief Funct Genomic Proteomic. 2009;8:136–44. doi: 10.1093/bfgp/eln055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rachdaoui N, Austin L, Kramer E, Previs MJ, Anderson VE, Kasumov T, et al. Measuring proteome dynamics in vivo: as easy as adding water? Mol Cell Proteomics. 2009;8:2653–63. doi: 10.1074/mcp.M900026-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nobeli I, Ponstingl H, Krissinel EB, Thornton JM. A structure-based anatomy of the E-coli metabolome. J Mol Biol. 2003;334:697–719. doi: 10.1016/j.jmb.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 28.Sekiguchi Y, Mitsuhashi N, Kokaji T, Miyakoda H, Mimura T. Development of a comprehensive analytical method for phosphate metabolites in plants by ion chromatography coupled with tandem mass spectrometry. J Chromatogr A. 2005;1085:131–6. doi: 10.1016/j.chroma.2005.01.098. [DOI] [PubMed] [Google Scholar]
- 29.Nemutlu E, Zhang S, Gupta A, Juranic NO, Macura SI, Terzic A, et al. Dynamic phosphometabolomic profiling of human tissues and transgenic models by O-18-assisted P-31 NMR and mass spectrometry. Physiol Genomics. 2012;44:386–402. doi: 10.1152/physiolgenomics.00152.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nemutlu E, Juranic N, Zhang S, Ward LE, Dutta T, Nair KS, et al. Electron spray ionization mass spectrometry and 2D P-31 NMR for monitoring O-18/O-16 isotope exchange and turnover rates of metabolic oligophosphates. Anal Bioanal Chem. 2012;403:697–706. doi: 10.1007/s00216-012-5899-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juranic N, Nemutlu E, Zhang S, Dzeja P, Terzic A, Macura S. 31P NMR correlation maps of 18O/16O chemical shift isotopic effects for phosphometabolite labeling studies. J Biomol NMR. 2011;50:237–45. doi: 10.1007/s10858-011-9515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villas-Boas SG, Bruheim P. The potential of metabolomics tools in Bioremediation studies. OMICS. 2007;11:305–13. doi: 10.1089/omi.2007.0005. [DOI] [PubMed] [Google Scholar]
- 33.Brown M, Dunn WB, Dobson P, Patel Y, Winder CL, Francis-McIntyre S, et al. Mass spectrometry tools and metabolite-specific databases for molecular identification in metabolomics. Analyst. 2009;134:1322–32. doi: 10.1039/b901179j. [DOI] [PubMed] [Google Scholar]
- 34.Pucar D, Dzeja PP, Bast P, Juranic N, Macura S, Terzic A. Cellular energetics in the preconditioned state – Protective role for phosphotransfer reactions captured by O-18-assisted P-31 NMR. J Biol Chem. 2001;276:44812–9. doi: 10.1074/jbc.M104425200. [DOI] [PubMed] [Google Scholar]
- 35.Zeleznikar RJ, Heyman RA, Graeff RM, Walseth TF, Dawis SM, Butz EA, et al. Evidence for compartmentalized adenylate kinase catalysis serving a high energy phosphoryl transfer function in rat skeletal muscle. J Biol Chem. 1990;265:300–11. [PubMed] [Google Scholar]
- 36.Knowles JR. Enzyme-catalyzed phosphoryl transfer reactions. Annu Rev Biochem. 1980;49:877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- 37.Weber DJ, Bhatnagar SK, Bullions LC, Bessman MJ, Mildvan AS. NMR and isotopic exchange studies of the site of bond cleavage in the MutT reaction. J Biol Chem. 1992;267:16939–42. [PubMed] [Google Scholar]
- 38.Dzeja P, Terzic A. Adenylate kinase and AMP signaling networks: metabolic monitoring, signal communication and body energy sensing. Int J Mol Sci. 2009;10:1729–72. doi: 10.3390/ijms10041729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dzeja PP, Bast P, Pucar D, Wieringa B, Terzic A. Defective metabolic signaling in adenylate kinase AK1 gene knock-out hearts compromises post-ischemic coronary reflow. J Biol Chem. 2007;282:31366–72. doi: 10.1074/jbc.M705268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dzeja PP, Hoyer K, Tian R, Zhang S, Nemutlu E, Spindler M, et al. Rearrangement of energetic and substrate utilization networks compensate for chronic myocardial creatine kinase deficiency. J Physiol. 2011;589:5193–211. doi: 10.1113/jphysiol.2011.212829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dzeja PP, Terzic A. Phosphotransfer reactions in the regulation of ATP-sensitive K+ channels. FASEB J. 1998;12:523–9. doi: 10.1096/fasebj.12.7.523. [DOI] [PubMed] [Google Scholar]
- 42.Dzeja PP, Redfield MM, Burnett JC, Terzic A. Failing energetics in failing hearts. Curr Cardiol Rep. 2000;2:212–7. doi: 10.1007/s11886-000-0071-9. [DOI] [PubMed] [Google Scholar]
- 43.Dzeja PP, Terzic A. Mitochondria-nucleus energetic communication: role for phosphotransfer networks in processing cellular information. In: Gibson G, Dienel G, editors. Handbook of neurochemistry and molecular neurobiology. New York (NY): Springer; 2007. p. 641-66. [Google Scholar]