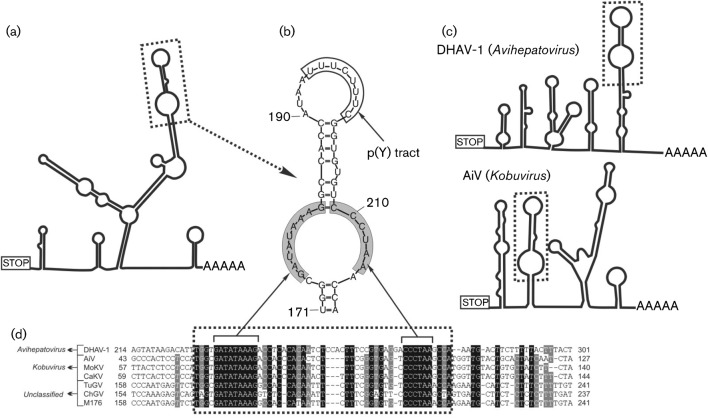
Fig. 3.
Conserved motif analysis of the 3′ UTR genome region. (a, c) The predicted secondary 3′ UTR structures of turkey/M176/2011/HUN (a) and of representative members of the genera Avihepatovirus and Kobuvirus (c). Dotted boxes indicate the conserved ‘barbell-like’ structure. (b) Detailed structure of the ‘barbell-like’ formation of the study sequence 3′ UTR. Grey boxes indicate identical nucleotides of different picornavirus species. (d) Sequence alignment of partial 3′ UTRs including the ‘barbell-like’ structure (dotted box) of duck hepatitis A virus 1 (DHAV-1; GenBank accession no. DQ249299), Aichi virus (AiV; FJ890523), mouse kobuvirus (MoKV; JF755427), canine kobuvirus (CaKV; JN088541), turkey gallivirus (TuGV; JF424829), chicken gallivirus (ChGV; JF424824) and the study sequence M176 (turkey/M176/2011/HUN; JQ691613). Note that the numbering of the 3′ UTR alignment begins from the STOP codon.