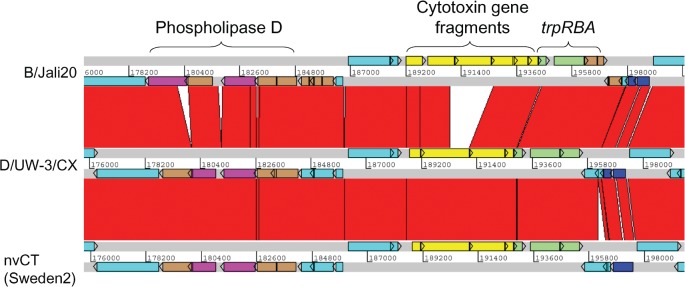
Fig. 2.
Comparison of the PZ in C. trachomatis nvCT (Sweden2, serovar E), Jali20 (serovar B) and D/UW-3/CX (serovar D). The grey lines indicate forward and reverse reading frames of sequenced genomes, with predicted CDSs superimposed. The red bars indicate regions of 95–100 % nucleotide identity. Brown CDSs denote pseudogenes. The phospholipase D locus (purple) contains pseudogenes in all strains. The cytotoxin locus (yellow) in D/UW-3/CX produces an active cytotoxin (Belland et al., 2001). This locus is identical in the nvCT, except for an SNP causing a putative 35 aa N-terminal truncation in the leftmost CDS. The trp operon (green) is complete in strains D/UW-3/CX and the nvCT, but has pseudogene trpA in Jali20. The nvCT carries an insertion of 307 bp in a CDS encoding a hypothetical protein downstream of the trp operon, causing an N-terminal extension of 127 aa. Additionally, an SNP in the nvCT relative to D/UW-3/CX causes read-through of CDSs encoding two conserved hypothetical proteins to create one CDS of 247 aa (dark blue). Other CDSs are turquoise.